Abstract
Members of the genus
Acknowledgements
This study was supported by a grant from the National Institute of Biological Resources (NIBR), funded by of the Ministry of Environment (MOE) and the National Research Foundation of Korea (NRF), funded by the Ministry of Science, ICT & Future Planning (2016R1C1B2008013) , Republic of Korea.
Figures & Tables
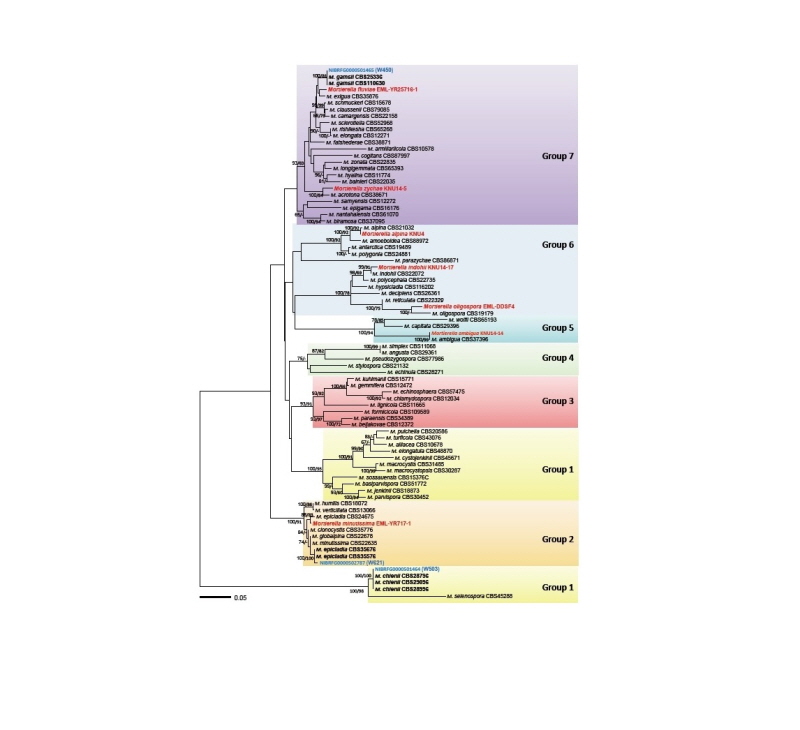
Fig. 1. Minimum evolution tree based on the internal transcribed spacer rDNA sequences. Bootstrapping support values of minimum evolution and maximum likelihood methods higher than 60% are given above or below the branches. The species revealed in the present study are shown in blue while the previously known species are shown in red. The scale bar equals the number of nucleotide substitutions per site.


