Introduction
Fungi belonging to the genus Tuber F. H. Wigg. (Pezizales, Ascomycota) form a cleistothecial ascomata underground, which is commonly known as “Truffle” [1]. They have ectomycorrhizal (ECM) relationships with host plants [2]. Tuber species are widely distributed in the forests of the north temperate zone, including those in China and Japan [1]. Some of these have a particular aroma, owing to which they have high commercial value [1]. However, only a few reports have been published on Tuber spp. in Korea, which have primarily focused on the morphological characteristics of the ascoma [3] or the DNA sequences from the ECM roots without the fungi [4,5]. This is the first study to report the collection of the fruiting bodies and ECM root tips of Tuber huidongense Y. Wang in Korea, along with a description of its morphological characteristics and phylogenetic analyses.
The fruiting bodies were collected from the rhizosphere of Quercus dentata Thunb in Pohang, Korea (N35°57'16″, E129°31'20″). The morphological characteristics of the fruiting bodies were evaluated, and genomic DNA was extracted from the ascoma using a DNeasy Plant Mini Kit (Qiagen GmvH, Hilden, Germany). For molecular identification, the ITS1F and ITS4 primers [6] were used to amplify the internal transcribed spacer (ITS) region, and the LR0R and LR16 primers [7] were used to amplify the large subunit (LSU) DNA. Nested PCR was performed to amplify the β-tubulin (TUB) DNA [8]. The first PCR experiment was performed using the Bt2a and Bt2b primers [9] for amplifying β-tubulin (TUB) DNA, in which the annealing temperature was set to 55℃. The first PCR product was diluted (1:10) and used as a template for the second PCR, which was performed using the Tuber-specific primers, Tubtubf and Elytubr [8]. The annealing temperature was set to 63℃, and DNA sequence analysis was performed using the final round PCR products (SolGent, Daejeon, Korea). The sequences were determined using BLAST (available from the website of the National Center for Biological Information, NCBI), and the phylogenetic tree was constructed using the neighbor-joining method with MEGA7 [10]. The ECM roots of the oak tree were also collected, and their morphological characteristics were studied. The ECM root tip samples were identified based on the DNA sequences of the (ITS) region amplified using the ITS1F and ITS4 primers [6].
Tuber huidongense Y. Wang, Mycotaxon 83: 191 (2002) [MB#383644]
Korean name: Huidong-deongi-beoseot (후이동덩이버섯); etymology from the species epithet derived from the name of the place where this species was first discovered
Classification: Ascomycota, Pezizales, Tuberaceae, Tuber
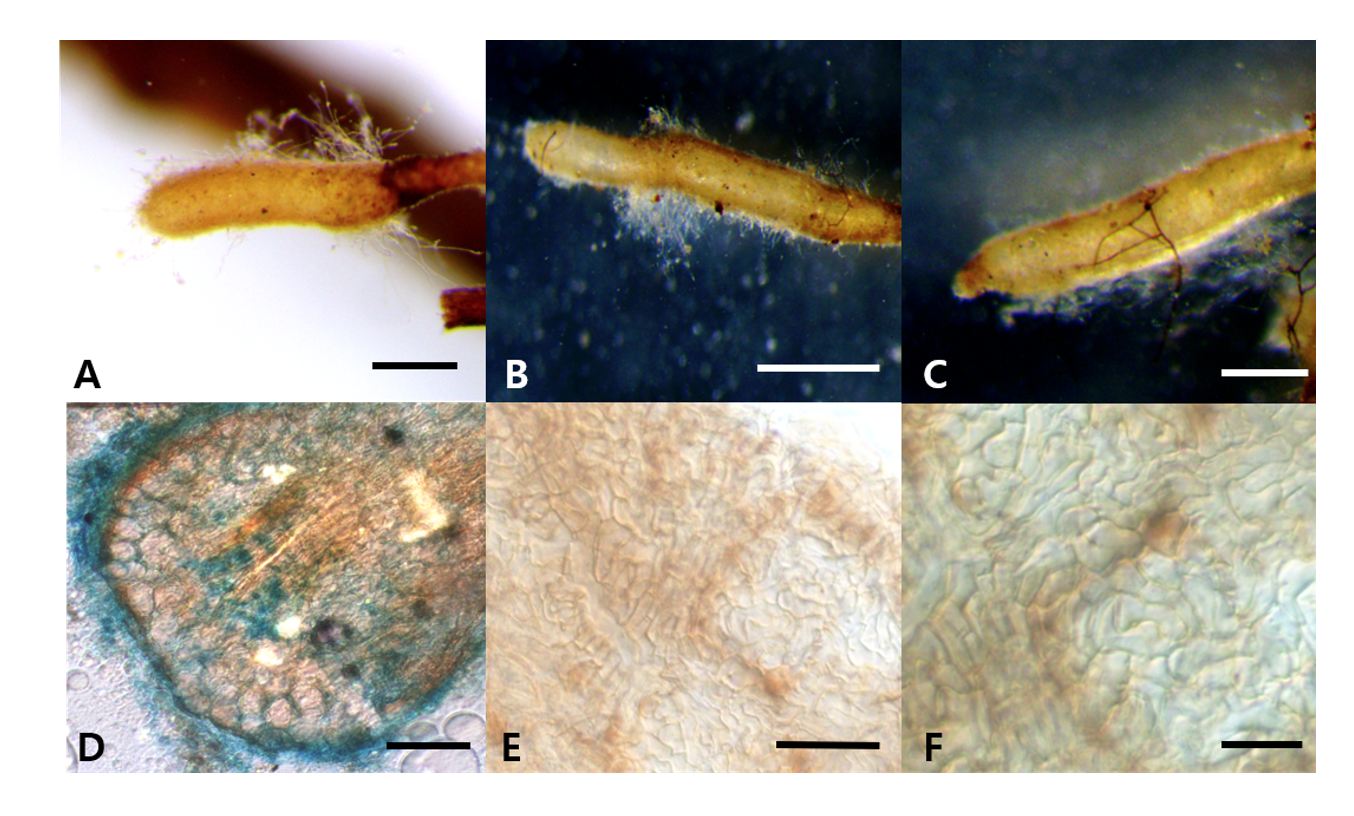
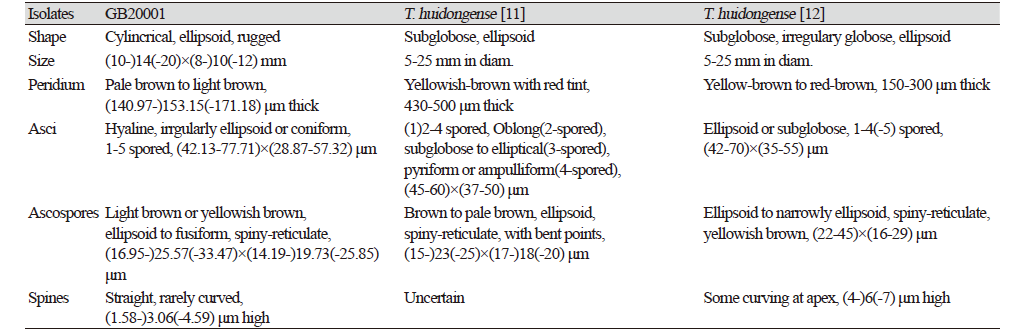
Morphology of ascoma: Cylindrical, short, elliptical, rugged, (10-)14(-20)×(8-)10(-12) mm in diam (Figs. 1A-C); Peridium: light or pale brown, approximately 140.97-153.15(-171.18) μm in thickness. Gleba: yellowish brown, paler than the peridium, white mycelia mixed in specific parts (Fig. 1D). Asci: hyaline, irregular, oval, or conical with rounded ends (Figs. 1E and 1F); 1-5 ascospores in each ascus, differing based on the number of ascospores;(42.13-)57.20(-77.71)×(28.87-)43.10(-57.32) μm Ascospore: light brown or yellowish brown; ellipsoid or fusiform; (16.95-)25.5(-33.47)×(14.19-)19.73(-25.85) μm (Fig. 1G). The reticular structure on the surface of the mature ascospore was similar to a turtle shell, with several spines present on the external surface; spines: (1.58-)3.06(-4.59) μm (Fig. 1H).
Specimen examined: Mt. Gwangjeongsan, Pohang-si, Gyeongsangbuk-do, Korea, N35°57'16″, E129°31'20", August 3, 2020, Tuber huidongense GB20001, collected from the rhizosphere of Quercus dentata; GenBank accession No. MT940543 (ITS rDNA from Ascoma), MT940570 (LSU), MT950763 (TUB), and MW199065 (ITS rDNA from ECM root).
Phylogenetic analysis: BLAST results showed that the ITS sequence of GB20001 was closely related to that of T. huidongense FJ797884 (98.75% similarity), the LSU gene sequence was closely related to that of T. huidongense GU979099 (99.51% similarity), and the TUB gene sequence was closely related to that of T. huidongense GU979149.1 (99.41% similarity). The phylogenetic tree constructed using the ITS, LSU, and TUB sequences confirmed that the DNA sequences of GB20001 belonged to the same monophyletic group as those of T. huidongense (Fig. 2). Therefore, the phylogenetic results validated the identification of GB20001 as T. huidongense.
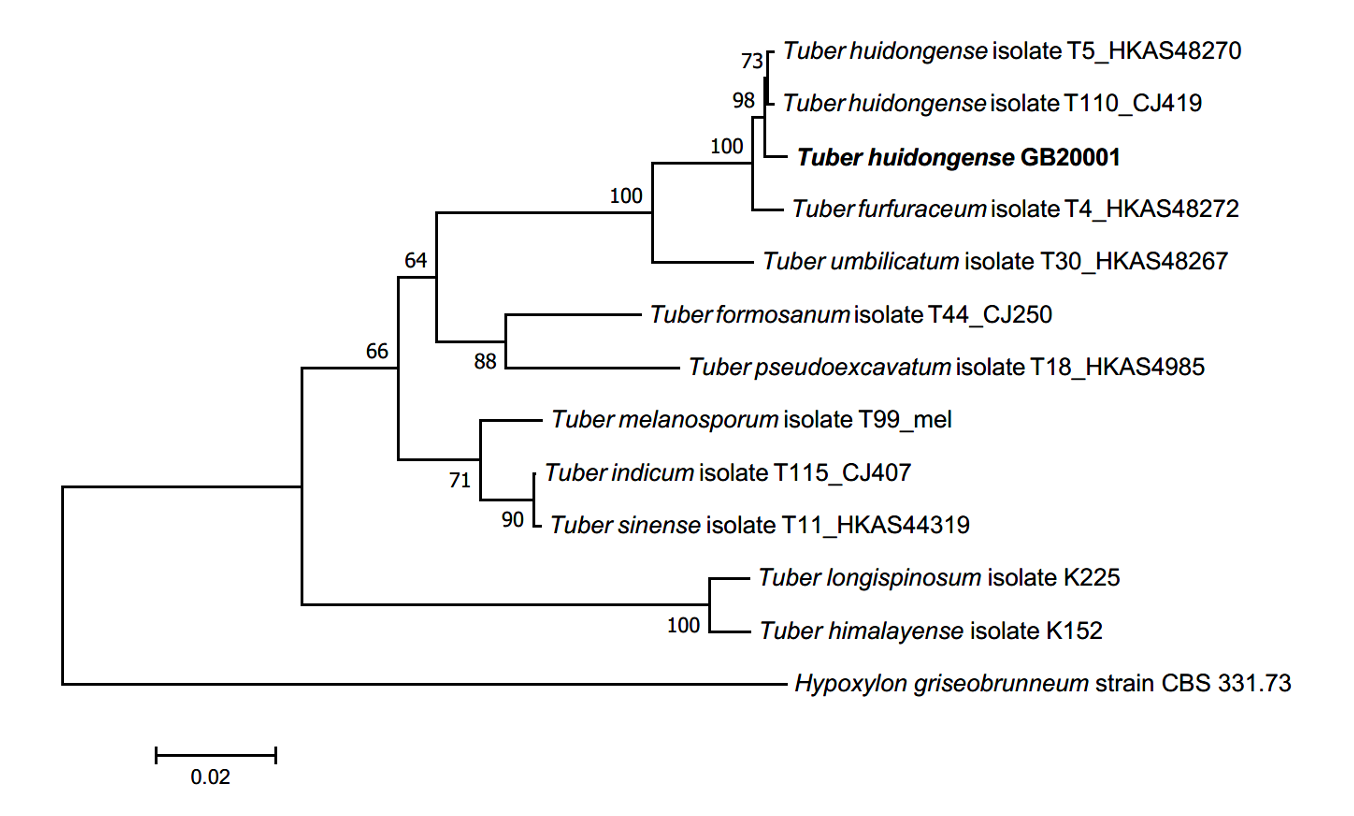
Fig. 2.Neighbor-joining phylogenetic tree of T. huidongense GB20001 ascoma based on concatenated alignment of internal transcribed spacer (ITS), large subunit (LSU) rDNA, and β-tubulin (TUB) DNA sequences. Hypoxylon griseobrunneum was used as an outgroup. Numbers on branches indicate bootstrap values (1,000 replicated).
Morphology of ECM roots: The ECM root tips were straight and cylindrical; their color varied from white to bright ivory; and white, fluffy emanating hyphae were observed on the surface (Figs. 3A-C). The cross section of the ECM showed the fungal mantle and Hartig net (Figs. 3D and 3E). The fungal mantle layer had a mosaic structure with irregularly shaped hyphae (Fig. 3F). The ECM root tips were identified as those of T. huidongense based on the DNA sequences of the ITS region (Fig. 4).
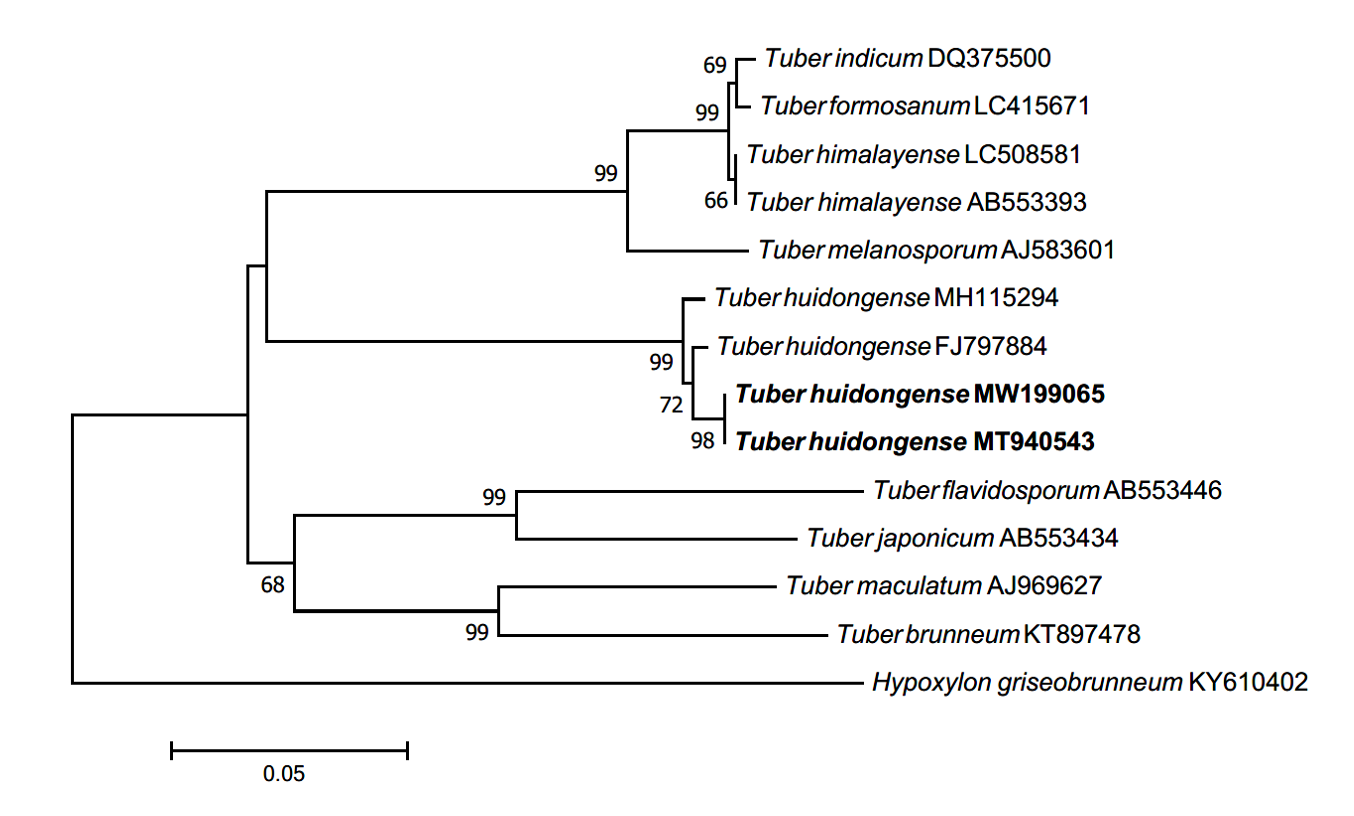
Fig. 4.Neighbor-joining phylogenetic tree of T. huidongense based on the alignment of internal transcribed spacer (ITS) rDNA sequences obtained from ectomycorrhizal root tips (MW199065) and ascoma of GB20001 (MT940543) in this study. Hypoxylon griseobrunneum was used as an outgroup. Numbers on branches indicate bootstrap values (1,000 replicates).
Wang and He [11] first reported the collection of T. huidongense in China; however, the morphological characterization and phylogeny were not clearly recorded, and there was no description of the ECM of T. huidongense. Deng et al. [12] reported additional morphological characteristics of T. huidongense and ectomycorrhiza formation with Pinus armandii. In this study, the ascoma and ascospores of GB20001 were found to be morphologically consistent with those of T. huidongense, which have been described in previous studies (Table 1). In addition, Deng et al. [12] reported that the ectomycorrhiza of T. huidongense with P. armandii. had a bifurcate morphotype, while ectomycorrhiza with different host, Q. dentata, observed in this study, had an unbranched morphotype (Fig. 3). In conclusion, to our knowledge, this is the first report on T. huidongense in Korea.