INTRODUCTION
The Ascomycetes is the largest phylum encompassing more than 33,000 species and a vast number of undescribed fungi. Information on the diversity and ecology of Korean fungal species belonging to the Ascomycetes (especially Dothideomycetes and Sordariomycetes) is still lacking. Dothideomycetes, previously known as Loculoascomycetes [1,2] is the largest group of ascomycetes with more than 23 orders, 119 families, 1,261 genera, and 19,000 species [3]. This group includes ecologically diverse fungi often found as plant pathogens, as saprobes degrading cellulose and other complex carbohydrates in dead or partially digested plant matters, in leaf litter or dung, as endophytes or epiphytes on living plants, some as coprophilous species, and a few as lichen-forming fungi [4]. They occur in almost every part of the world ranging from terrestrial to freshwater and marine habitats. Members of Dothideomycetes are characterized morphologically by ascolocular ascoma development and bitunicate, fissitunicate asci [1,5-8]. As regards to Sordariomycetes, this class is the second largest and comprises of 6 subclasses, 28 orders, 90 families, and 1,344 genera [9,10]. Species of Sordariomycetes have a cosmopolitan distribution, accommodates mostly terrestrial taxa as endophytes, saprobes, epiphytes, plant pathogens, fungicolous, lichenized or lichenicolous fungi and aquatic taxa as pathogens of arthropods and mammals [9-13]. Members of Sordariomycetes are mainly characterized by non-lichenized, perithecial ascomata and inoperculate unitunicate or non-fissitunicate asci [14,15]. Dothideomycetes and Sordariomycetes species have the capacity to produce an extensive array of secondary metabolites across the fungal kingdom [16,17]. Some of the secondary metabolites act as potential biocontrol agents and also exploited for various medicinal and industrial uses [18-21] whereas some are phytotoxic or mycotoxic [22].
The fungal taxa belonging to Dothideomycetes and Sordariomycetes have been documented well in Thailand, UK, China, India, and Japan [23,24-26]. The knowledge on the ecology of the species belonging to Dothideomycetes and Sordariomycetes are limited in Korea despite their role in ecosystem health, global carbon cycling as saprotrophs and degraders of plant biomass and metabolic profiles [27-29]. Therefore, the objective of this study is to characterize the unrecorded nine fungal species in Korea using both morphological and molecular analyses: C. robusta, F. acetilereae, H. pedis, H. snookiorum, M. fusiformis, Met. pemphigi, P. crystallina, S. candida and V. citrinella.
MATERIALS AND METHODS
Fungal strains isolation
The information related to sample sources, location and sampling sites are listed in Table 1. A dilution plating method was used for the isolation of fungal strains from freshwater, soil, and seawater. For isolating fungi from plant samples, a previously described methods was employed [30]. Pure isolates were transferred to new fresh potato dextrose Agar (PDA; Difco, Sparks, MD, USA) and were incubated for 4-5 days at 25℃. For stock storage, pure isolates were maintained in PDA slant tubes and 20% glycerol at -80℃ in the Environmental Microbiology Laboratory Fungarium, Chonnam National University, Gwangju, Korea as CNUFC BCSM3, CNUFC F7-24-5, CNUFC HRG1, CNUFC HRW1-12, CNUFC YJS7, CNUFC AS1-26, CNUFC PLTFB118, CNUFC KU1-1, and CNUFC DYR1-2. CNUFC BCSM3, CNUFC AS1-26, CNUFC PLTFB118, CNUFC KU1-1, CNUFC F7-24-5, CNUFC HRG1, CNUFC HRW1-12, CNUFC YJS7, and CNUFC DYR1-2, were also deposited at the Collection of National Institute of Biological Resources (NIBR) Incheon, Korea as NIBRFG0000503047, IMYKFGC000000029, NIBRFG0000503054, NIBRFGC000508425 and Nakdonggang National Institute of Biological Resources (NNIBR), Sangju, Korea as NNIBRFG9312, NNIBRFG31611, NNIBRFG31612, NNIBRFG9315 and NNIBRFG9334, respectively.
Morphological analysis
All the isolated strains were grown on PDA, malt extract agar (MEA; malt extract, 20 g; agar, 20 g; distilled water, 1 L), oatmeal agar (OA; oatmeal flakes, 30 g; agar, 20 g; distilled water, 1 L), and cornmeal agar (cornmeal 30 g; agar, 15 g; distilled water, 1 L) at 25℃ for morphological characterization. Samples were examined using an Olympus BX51 microscope with DIC optics (Olympus, Tokyo, Japan) by mounting in a lactophenol solution (Junsei Chemical Co., Ltd., Tokyo, Japan).
DNA extraction, PCR and sequencing
Fungal genomic DNA was extracted using a Solg Genomic DNA preparation Kit (Solgent Co., Ltd., Daejeon, Korea). Amplification of fragments of the gene regions of the large subunit of the nuclear ribosomal DNA (LSU), the internal transcribed spacers (ITS) and beta-tubulin (TUB) were performed using the following primer pairs LR0R/LR5 [31], ITS5/ITS4 [32], ITS5/LR7 [31,32], ITS1/NL4 [32,33] and T10/Bt2b [34]. PCR amplification was performed according to the conditions described by Hongsanan et al. [8], Luo et al. [12], and Sandoval-Denis et al. [35]. After amplification, PCR products were purified using an Accuprep PCR Purification Kit (Bioneer Corp., Daejeon, Korea). Sequencing was performed using the same primers as those used for amplification with an ABI PRISM 3730XL Genetic Analyzer (Applied Biosystems, Foster City, CA, USA).
Phylogenetic analysis
A homology search of DNA sequences was performed using the BLASTn algorithm of the National Center for Biotechnology Information (https://www.ncbi.nlm.nih.gov). Each strains sequences were aligned with reference sequences downloaded from GenBank using Clustal X version 2.1 [36] and edited manually using Bioedit version 7.2.6.0 [37]. Maximum likelihood (ML) phylogenies were constructed using MEGA version 7 [38].
ITS sequences of strains CNUFC BCSM3, CNUFC F7-24-5, CNUFC YJS7, CNUFC PLTFB118, CNUFC KU1-1, and CNUFC DYR1-2 were deposited in the GenBank database under accession numbers MZ435755, MZ435756, MZ435757, MZ435758, MZ435759 and MZ435760, respectively. LSU sequences of strains CNUFC BCSM3, CNUFC F7-24-5, CNUFC HRG1, CNUFC HRW1-12, CNUFC YJS7, CNUFC PLTFB118, and CNUFC DYR1-2 were deposited in the GenBank database under accession numbers MZ436990, MZ436991, MZ436992, MZ436993, MZ436994, OK021601 and MZ436995, respectively. TUB sequences of strains CNUFC AS1-26 and CNUFC KU1-1 were deposited in the GenBank database under accession numbers MZ450120 and MZ450121, respectively.
RESULTS
Phylogenetic analysis
According to the BLASTn, ITS sequences of CNUFC BCSM3, CNUFC F7-24-5, CNUFC YJS7, CNUFC PLTFB118, CNUFC KU1-1, and CNUFC DYR1-2 showed similarities of 100% (508/508 bp), 99.9% (1,045/1,046 bp), 98.7% (1,002/1,015 bp), 99.3% (1,052/1,060 bp), 99.8% (567/568 bp), and 100% (508/508 bp) with those of C. robusta CBS 508.84 (MH861773), F. acetilerea RF3 (LC333211), M. elegans NRRL 62999 (KM056317), P. crystallina CBS 681.95 (EU167579), S. candida CBS 389.52 (KX924012) and V. citrinella VC-101 (MK357063), respectively. Similarly, BLASTn using LSU sequences of CNUFC BCSM3, CNUFC F7-24-5, CNUFC HRG1, CNUFC HRW1-12, CNUFC YJS7, CNUFC PLTFB118, and CNUFC DYR1-2, showed similarities of 99.1% (541/546 bp), 99.3% (554/558 bp), 99.6% (512/514 bp), 99.5% (582/585 bp), 99.8% (550/551 bp), 99.7% (565/567 bp), and 99.8% (566/567 bp) with those of C. robusta CBS 508.84 (MH873473), F. siamensis MFLUCC 17-2577 (MT215550), H. pedis HKU35 (NG_056287), H. snookiorum ILLS00125755 (MH161189), M. fusiformis LC1701 (KX986140), P. crystallina ZJUM 2 (KP895884) and V. citrinella DAOM 226716 (HQ843770), respectively. BLASTn using TUB sequence of CNUFC AS1-26 and CNUFC KU1-1, showed similarities of 100% (614/614 bp) and 99.60% (493/495 bp) with Met. pemphigi ARSEF 7491 (KJ398591) and S. candida CBS 205.27 (KX924444), respectively. Phylogram generated from the Maximum Likelihood analysis for ITS, LSU, and TUB revealed that the strains, CNUFC BCSM3, CNUFC F7-24-5, CNUFC HRG1, CNUFC HRW1-12, CNUFC YJS7, CNUFC AS1-26, CNUFC PLTFB118, CNUFC KU1-1 and CNUFC DYR1-2 were identical to C. robusta, F. acetilerea, H. pedis, H. snookiorum, M. fusiformis, Met. pemphigi, P. crystallina, S. candida and V. citrinella, respectively (Figs. 1-8).
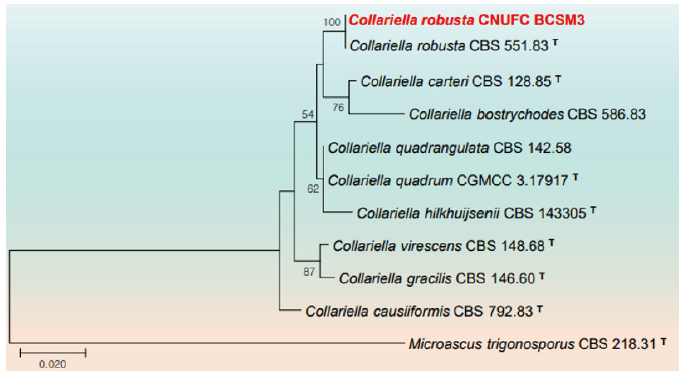
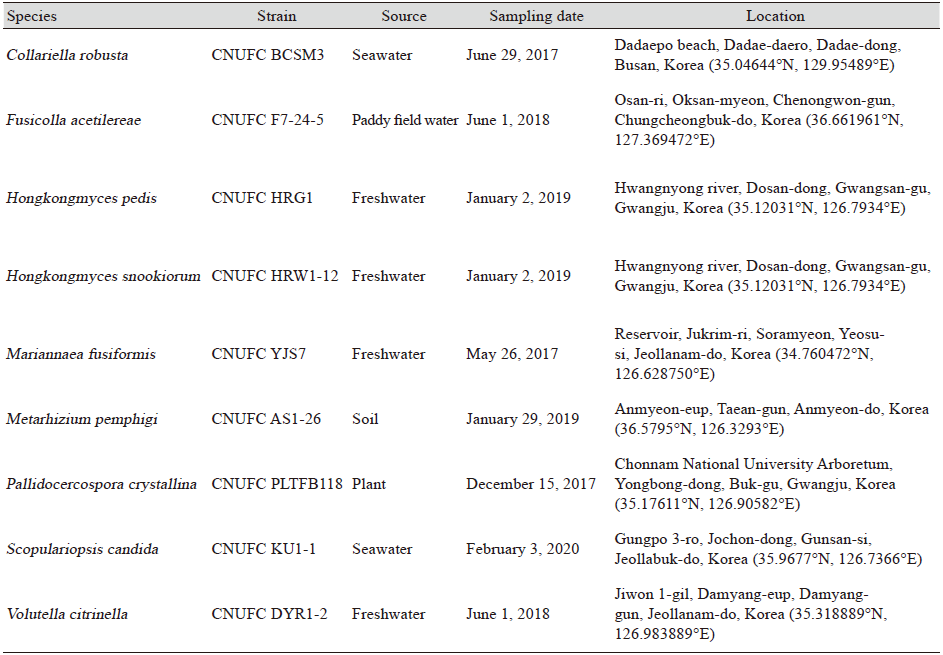
Fig. 1. Phylogenetic tree based on the maximum likelihood analyses of the combined internal transcribed spacers (ITS) and large subunit (LSU) rDNA sequences of Collariella robusta CNUFC BCSM3. Microascus trigonosporus CBS 218.31 was used as outgroup. Bootstrap support values of ≥50% from 1,000 replicates are indicated at the nodes. The bar indicates the number of substitutions per position. T=type.
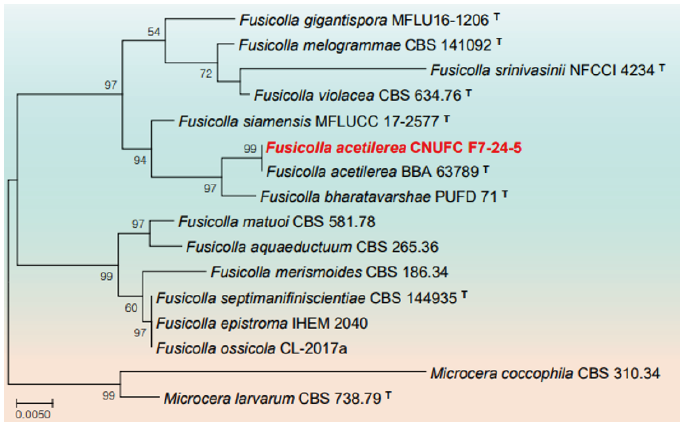
Fig. 2. Phylogenetic tree based on the maximum likelihood analysis of the combined internal transcribed spacers (ITS) and large subunit (LSU) rDNA sequences of Fusicolla acetilerea CNUFC F7-24-5. Microcera larvarum CBS 738.79 and M. coccophila CBS 310.34 were used as outgroups. Bootstrap support values of ≥50% from 1,000 replicates are indicated at the nodes. The bar indicates the number of substitutions per position. T=type.
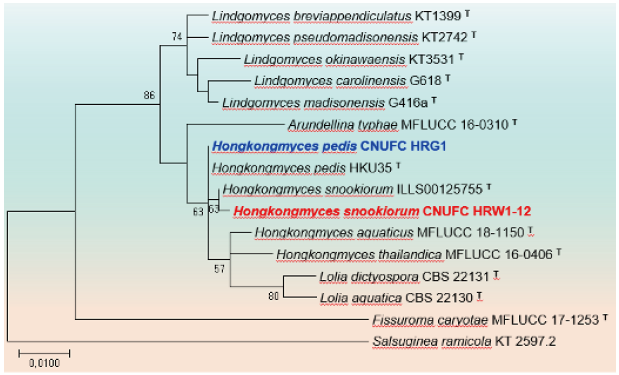
Fig. 3. Phylogenetic tree based on the maximum likelihood analysis of large subunit (LSU) rDNA sequences of Hongkongmyces pedis CNUFC HRG1 and Hongkongmyces snookiorum CNUFC HRW1-12. Salsuginea ramicola KT2597.2 was used as outgroup. Bootstrap support values of ≥50% from 1,000 replicates are indicated at the nodes. The bar indicates the number of substitutions per position. T=type.
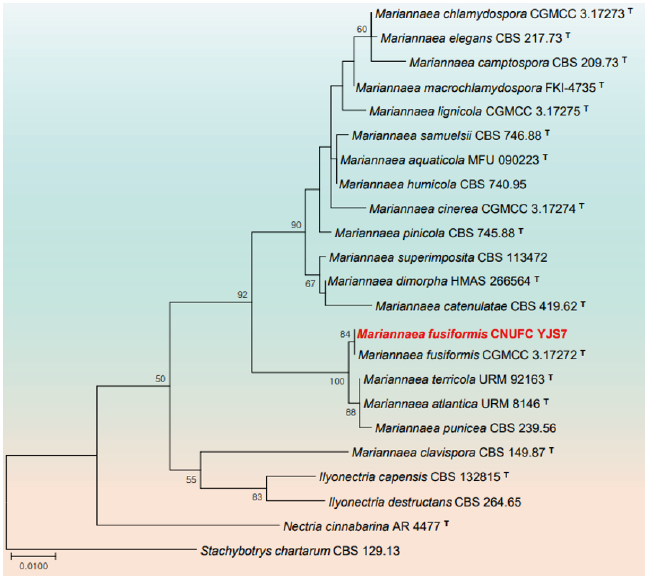
Fig. 4. Phylogenetic tree based on the maximum likelihood analysis of the combined internal transcribed spacers (ITS) and large subunit (LSU) rDNA sequences of Mariannaea fusiformis CNUFC YJS7. Stachybotrys chartarum CBS 129.13 was used as outgroup. Bootstrap support values of ≥50% from 1,000 replicates are indicated at the nodes. The bar indicates the number of substitutions per position. T=type
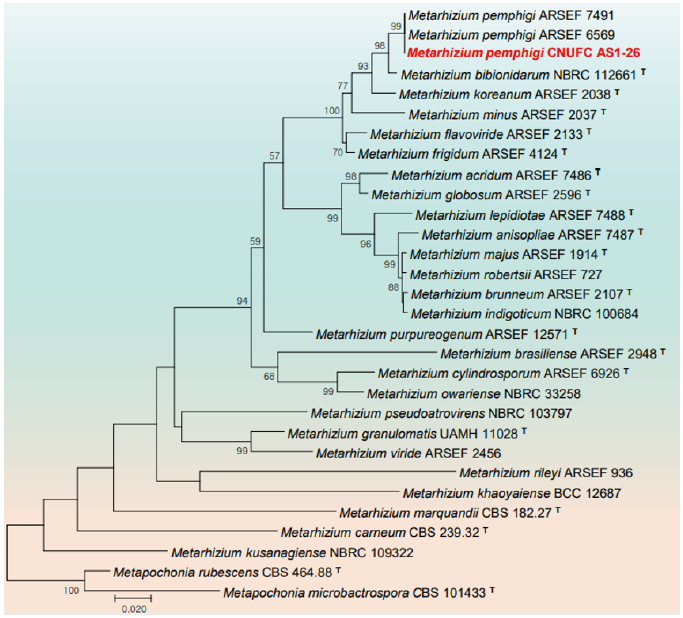
Fig. 5. Phylogenetic tree based on the maximum likelihood analysis of beta-tubulin (TUB) sequences of Metarhizium pemphigi CNUFC AS1-26. Metapochonia rubescens CBS 464.88 and Metapochonia microbactrospora CBS 101433 were used as outgroups. Bootstrap support values of ≥50% from 1,000 replicates are indicated at the nodes. The bar indicates the number of substitutions per position. T=type.
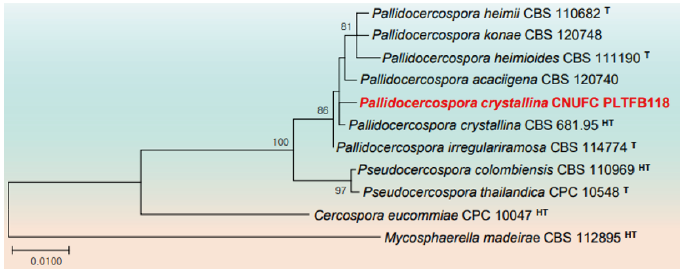
Fig. 6. Phylogenetic tree based on the maximum likelihood analysis of the combined internal transcribed spacers (ITS) and large subunit (LSU) rDNA sequences of Pallidocercospora crystallina CNUFC PLTFB118. Mycosphaerella madeirae CBS 112895 was used as outgroup. Bootstrap support values of ≥50% from 1,000 replicates are indicated at the nodes. The bar indicates the number of substitutions per position. T=type, HT=ex-holotype.
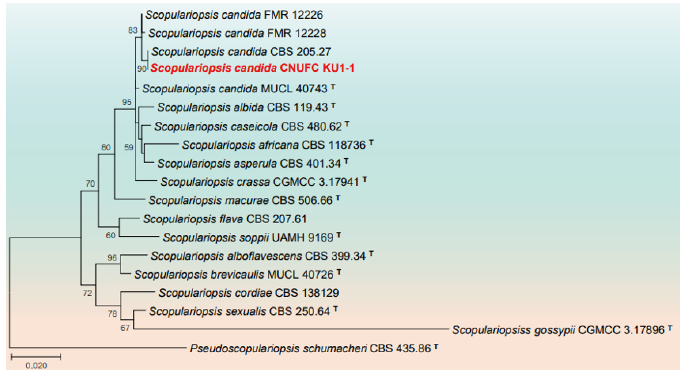
Fig. 7. Phylogenetic tree based on the maximum likelihood analysis of internal transcribed spacers (ITS) rDNA and beta-tubulin (TUB) sequences of Scopulariopsis candida CNUFC KU1-1. Pseudoscopulariopsis schumacheri CBS 435.86 was used as outgroup. Bootstrap support values of ≥50% from 1,000 replicates are indicated at the nodes. The bar indicates the number of substitutions per position. T=type.
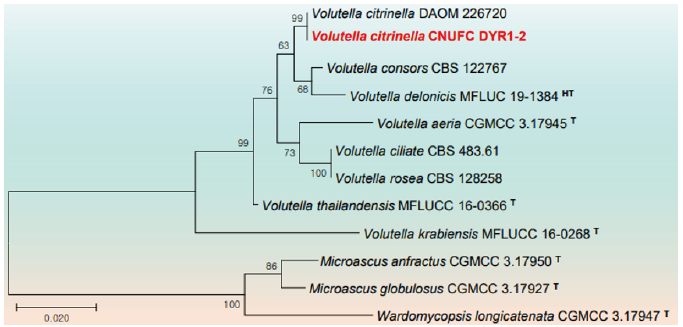
Fig. 8. Phylogenetic tree based on the maximum likelihood analysis of the combined internal transcribed spacers (ITS) and large subunit (LSU) rDNA sequences of Volutella citrinella CNUFC DYR1-2. Wardomycopsis longicatenata CGMCC 3.17947, Microascus anfractus CGMCC 3.17950, and M. globulosus CGMCC 3.17927 were used as outgroups. Bootstrap support values of ≥50% from 1,000 replicates are indicated at the nodes. The bar indicates the number of substitutions per position. T=type, HT=ex-holotype.
Taxonomy
Collariella robusta (L.M. Ames) X. Wei Wang & Samson, Studies in Mycology 84: 217 (2016) (Fig. 9).
≡Chaetomium robustum L.M. Ames, A monograph of the Chaetomiaceae: 35 (1963).
≡Chaetomium caprinum Bainier, Bulletin de la Société Mycologique de France 25 (3): 223 (1910).

Fig. 9. Morphology of Collariella robusta CNUFC BCSM3. (A, D) Colonies on potato dextrose agar (PDA). (B, E) Colonies on malt extract agar (MEA). (C, F) Colonies on oatmeal agar (OA) (A-C: Obverse view, D-F: Reverse view). (G, H) Mature ascomata. (I) Asci. (J) Ascospores (scale bars: G, H=20 μm, I, J=10 μm).
Description: Colonies on PDA plane, regular, cottony, white, reverse white, measuring 24-28 mm in diameter after 7 d at 25℃. Colonies on MEA plane, regular, transparent white, reverse transparent white, measuring 23-26 mm after 7 d at 25℃. Colonies on OA regular, white, reverse pale white, measuring 23-27 mm after 7 d at 25℃.
Micromorphology: Ascomata were ampulliform, elongated and measuring 260-410 × 160-303 μm. Asci were club-shaped, 8-spored and measuring 32-37 × 8-11 μm. Ascospores were limoniform and measuring 5.9-6.5 × 4.7-5.6 μm.
Remarks: Species of Collariella is morphologically characterized by the production of a dark collar-like apex around the ostiolar pore of the ascomata [39]. CNUFC BCSM3 shares similar morphological features to C. robusta [39]. Also, the phylogeny based on combined ITS and LSU rDNA placed CNUFC BCSM3 with C. robusta CBS 551.83 (T). Collariella robusta was reported to occur in litter and woodlot soil from Jamaica and in rabbit pellets from the United Kingdom [39,40]. The species was isolated from seawater providing further information regarding the ecological characteristics of this species.
Fusicolla acetilerea (Tubaki, C. Booth & T. Harada) Gräfenhan & Seifert, Studies in Mycology 68: 100 (2011) (Fig. 10).
≡Fusarium merismoides var. acetilereum Tubaki, C. Booth & T. Harada, Transactions of the British Mycological Society 66 (2): 355 (1976).
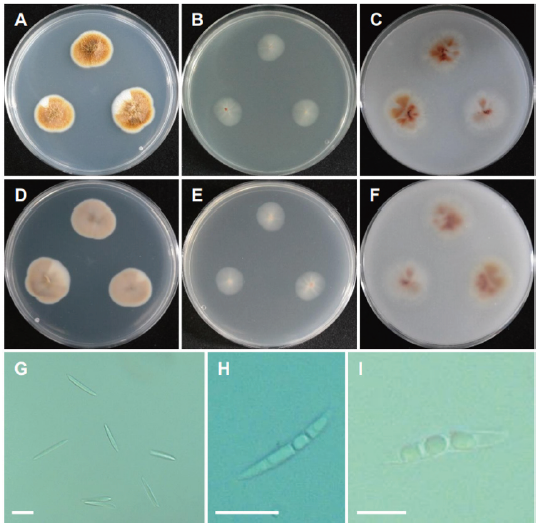
Fig. 10. Morphology of Collariella robusta CNUFC BCSM3. (A, D) Colonies on potato dextrose agar (PDA). (B, E) Colonies on malt extract agar (MEA). (C, F) Colonies on oatmeal agar (OA) (A-C: Obverse view, D-F: Reverse view). (G, H) Mature ascomata. (I) Asci. (J) Ascospores (scale bars: G, H=20 μm, I, J=10 μm).
Description: Colonies on PDA floccose, regular, pale yellow orange, reverse pale yellowish brown, measuring 22-24 mm after 7 d at 25℃. Colonies on MEA plane, regular, transparent white, reverse pale white, measuring 20-23 mm after 7 d at 25℃. Colonies on OA plane, regular, white mycelium to pale orange toward center, reverse white mycelium to pale orange, measuring 23-25 mm after 7 d at 25℃.
Micromorphology: Macroconidia were long, slender, falcate, with 3-4 transverse septa and measuring 40-58 × 2.5-4 μm. Chlamydospore was not observed. No perithecia were produced.
Remarks: The genus Fusicolla is characterized by superficial, nonstromatic, globose to sub-globose, pale yellow to pale orange ascomata, one-septate, ellipsoidal, hyaline and straight or slightly curved ascospores [41,42]. Fusicolla acetilerea was described with phialidic, straight or curved with a round apex and a foot cell macroconidia mostly with 3(-4) transverse septa when mature, measuring (30-)35-40 × 3.5(-4.0) μm [43]. However, CNUFC F7-24-5 isolate can be differentiated from previous described F. acetilerea by producing relatively longer macroconidia size. Fusicolla acetilerea was found to occur mostly in soil habitats, on decaying wood, and on the beetle Xyleborinus saxesenii [41,44,45]. CNUFC F7-24-5 is reported to occur in a freshwater environment for the first time.
Hongkongmyces pedis C.C. Tsang, J.F.W. Chan, N.J. Trendell-Smith, A.H.Y. Ngan, I.W.H. Ling, S.K.P. Lau & P.C.Y. Woo, Medical Mycology 52 (7): 740 (2014) (Fig. 11).
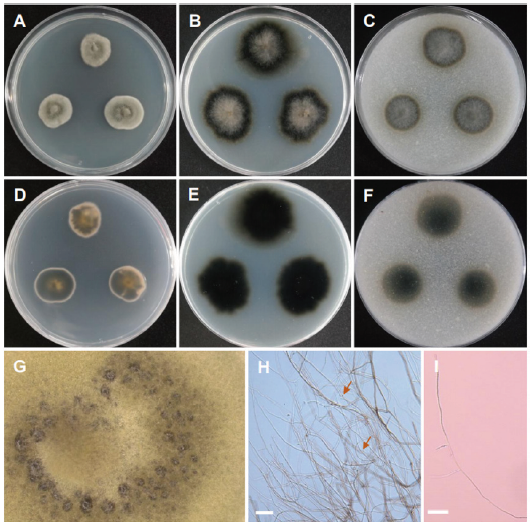
Fig. 11. Morphology of Hongkongmyces pedis CNUFC HRG1. (A, D) Colonies on potato dextrose agar (PDA). (B, E) Colonies on malt extract agar (MEA). (C, F) Colonies on cornmeal agar (CMA). (A-C: Obverse view, D-F: Reverse view). (G) Dark hyphae (observed under stereomicroscope). (H, I) Only sterile mycelia consisted of gray and septate hyphae consists of round and spore-like bodies shown by arrow (scale bars=20 μm).
Description: Colonies on PDA greyish green, slow growing, reverse slight yellow pigments, measuring 21-23 mm after 14 d at 25℃. Colonies on MEA slightly irregular, grey, velvety, reverse dark olive, measuring 26-27 mm after 14 d at 25℃. Colonies on CMA slightly regular, dark olive grey, reverse olive, measuring 23-26 mm after 14 d at 25℃.
Micromorphology:Only sterile mycelia were produced after 3 months of incubation on all three media. On PDA, hyphae become darkly pigmented and round, spore-like bodies occasionally present along the hyphae. Hyphae were septate, narrow, and branched. Mycelia were sterile. No fruiting bodies or conidia were produced.
Remarks: The genus Hongkongmyces was erected with H. pedis HKU35 (T) isolated from biopsy tissues of a patient’s infected foot as type species [46]. Morphologically H. pedis HKU35 (T) was characterized by forming grey colonies on media, composed of grey, narrow, septate, branched hyphae with acute angles, sterile mycelia with no fruiting bodies or sporulating structures. CNUFC HRG1 is morphologically similar to H. pedis HKU35 (T) with producing sterile mycelia and narrow, septate, branched hyphae. Similarly, CNUFC HRG1 clustered within the same clade with H. pedis HKU35 (T) in the phylogeny based on LSU rDNA.
Hongkongmyces snookiorum Raudabaugh, Iturr. & A.N. Mill., Persoonia 40: 289 (Fig. 12).
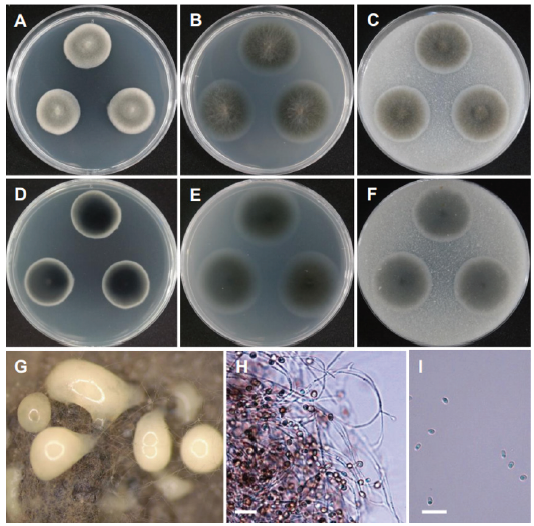
Fig. 12. Morphology of Hongkongmyces pedis CNUFC HRG1. (A, D) Colonies on potato dextrose agar (PDA). (B, E) Colonies on malt extract agar (MEA). (C, F) Colonies on cornmeal agar (CMA). (A-C: Obverse view, D-F: Reverse view). (G) Dark hyphae (observed under stereomicroscope). (H, I) Only sterile mycelia consisted of gray and septate hyphae consists of round and spore-like bodies shown by arrow (scale bars=20 μm).
Description: Colonies on PDA regular, dull greyish green, reverse dark olive green, measuring 23-25 mm after 14 d at 25℃. Colonies on MEA regular, hyaline, olive, reverse olive grey, measuring 26-28 mm after 14 d at 25℃. Colonies on CMA regular, pale olive, reverse olive grey, measuring 24-27 mm after 14 d at 25℃.
Micromorphology: Conidiogenous cells were discrete, hyaline, phialidic, subulate and tightly aggregated. Conidia were hyaline, ellipsoid to ovoid and measuring 4.3-5.3 × 3.2-4 μm.
Remarks: Hongkongmyces snookirorum is coelomycetous with globose to ampulliform pycnidia, hyaline, subulate to ampulliform conidiogenous cells with sympodial proliferations, hyaline, ellipsoid to ovoid conidia [47]. The species was also reported from freshwater and appeared to be identical with that previously isolated from detritus submerged in a freshwater fen [47].
Mariannaea fusiformis D.M. Hu & L. Cai, Mycological Progress 16: 278 (2017) (Fig. 13).
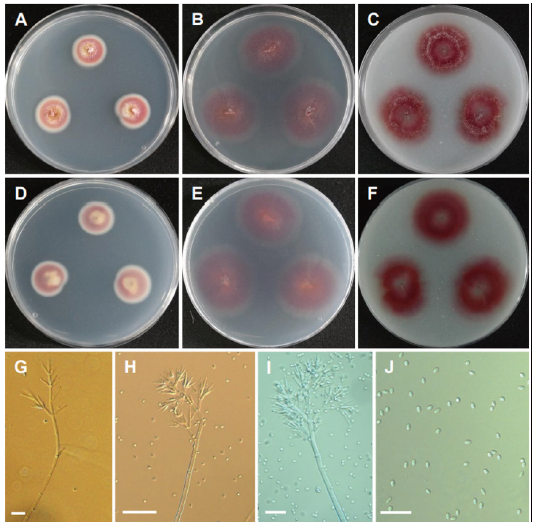
Fig. 13. Morphology of Mariannaea fusiformis CNUFC YJS7. (A, D) Colonies on potato dextrose agar (PDA). (B, E) Colonies on malt extract agar (MEA). (C, F) Colonies on oatmeal agar (OA). (A-C: Obverse view, D-F: Reverse view). (G-I) Conidiophores and verticillate branches of phialides. (J) Conidia (scale bars: G, I, J=20 μm, H= 50 μm).
Description: Colonies on PDA floccose, zonate, white to purple and yellowish at the center, reverse purple and yellowish at the center, measuring 22-23 mm after 7 d at 25℃. Colonies on MEA plane, regular, pale purple, reverse pale purple, measuring 26-28 mm after 7 d at 25℃. Colonies on OA plane, regular, purple, light cottony mycelium, reverse pale purple, measuring 24-26 mm after 7 d at 25℃.
Micromorphology: Conidiophores were erect, septate, cylindrical, macronematous, mononematous and measuring 550-800 μm long. Phialides were hyaline, smooth-walled, ampulliform and measuring 16.9-35.8 × 4-5 μm. Conidia were fusiform to subglobose, hyaline, smooth, and aseptate, produced in imbricate chains and measuring 4.9-7 × 3-4 μm. Chlamydospores were hyaline, thick-walled, intercalary or terminal and measuring 6.6-9.2 × 4.9-6.5 μm.
Remarks: The genus Mariannaea is characterized by flask-shaped phialides, distinctly stalked and one- or two-celled conidia forming imbricate or straight chains [48]. Mariannaea fusiformis CGMCC 3.17272 (T) isolated from submerged wood is characterized by its purple colonies on PDA, fusiform to subglobose conidia [49]. CNUFC YJS7 also shows purple colonies cultured on PDA, MEA and OA media and fusiform to subglobose conidia. CNUFC YJS7 is reported from freshwater. The species was isolated from freshwater showing that the species are associated with freshwater habitat.
Metarhizium pemphigi (Driver & Milner) Kepler, S.A. Rehner & Humber, Mycologia 106: 824 (2014) (Fig. 14).
≡Metarhizium flavoviride var. pemphigi Driver & Milner, Mycological Research 104 (2): 144 (2000).
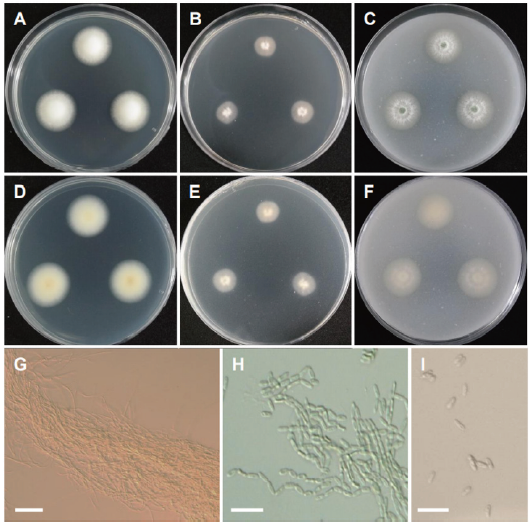
Fig. 14. Morphology of Metarhizium pemphigi CNUFC AS1-26. (A, D) Colonies on potato dextrose agar (PDA). (B, E) Colonies on malt extract agar (MEA). (C, F) Colonies on oatmeal agar (OA). (A-C: Obverse view, D-F: Reverse view). (G, H) Basipetal chains comprising conidia and conidiophores forming a sporulating layer as conidial columns. (I) Conidia (scale bars: G=10 μm, H, I=20 μm).
Description: Colonies on PDA floccose, cottony white, reverse pale white, measuring 24-26 mm after 7 d at 25℃. Colonies on MEA plane, regular, pale orange, white cottony towards the center, reverse pale orange, measuring 18-20 mm after 7 d at 25℃. Colonies on OA plane, regular, white to light green, reverse pale orange, measuring 25-27 mm after 7 d at 25℃.
Micromorphology: Conidiophores formed a sporulating layer as conidial columns; basipetal chains were formed in conidiation. Conidia were uninucleate and ovoid to cylindrical and measuring 4.8-7.4 × 2.0-2.3 μm.
Remarks: The genus Metarhizium is characterized by the production of conidia in long chains, dense phialides in parallel arrangement, and aggregated conidiophores with repeated, verticillate branching [50,51]. Metarhizium pemphigi is characterized by ovoid to elongate conidia, measuring 5.4 (±0.47)×2.4 (±0.43) μm, occasionally <9 μm long, borne in chains on cylindrical phialides and light green spore [52]. Metarhizium pemphigi was previously isolated from forest or wood soil, Pemphigus treherni, other Hemiptera, and Melanotus cribricollis [52-55]. Our isolate CNUFC AS1-26 was isolated from soil environment.
Pallidocercospora crystallina (Crous & M.J. Wingf.) Crous & M.J. Wingf., Studies in Mycology 75: 74 (2012) (Fig. 15).
≡Pseudocercospora crystallina Crous & M.J. Wingf., Mycologia 88 (3): 451 (1996).
≡Mycosphaerella crystallina Crous & M.J. Wingf., Mycologia 88 (3): 451 (1996).

Fig. 15. Morphology of Pallidocercospora crystallina CNUFC PLTFB118. (A, D) Colonies on potato dextrose agar (PDA). (B, E) Colonies on malt extract agar (MEA). (C, F) Colonies on oatmeal agar (OA). (A-C: Obverse view, D-F: Reverse view). (G) Plant sample. (H, I) Dark hyphae (observed under stereomicroscope). (J, K) Hyphae (scale bars=20 μm).
Description: Colonies on PDA velvety, olive green, radially concentrated toward the center, reverse iron-grey, measuring 23-26 mm after 7 d at 25℃. Colonies on MEA regular, plane, peanut brown, reverse olive green, measuring 22-24 mm after 7 d at 25℃. Colonies on CMA regular, olive green, reverse dark olive, measuring 19-21 mm after 7 d at 25℃.
Micromorphology: Filamentous septate hyphae showed pale brown, thickened walls; no sporulation were observed.
Remarks: The genus Pallidocercospora was introduced to include Cercospora-like species however not congeneric with Cercospora and is typified by P. heimii Crous (termed Pseudocercospora heimii Crous) [56]. Previously, Pallidocercospora was believed to contain Mycosphaerella heimii complex species based on culture characteristics, with pale brown cercosporoid conidia, and segregated from Pseudocercospora species mainly by the distinct phylogenetic positions of its members. CNUFC PLTFB118 shows similar morphology to P. crystallina reported to cause infections in humans [57]. Similarly, Huang et al. [58] also reported that P. crystallina isolated from spot leaves of citrus cannot be induced to sporulate on artificial media. To the best of our knowledge, this is the first report of P. crystallina occurring as an endophyte in Korea.
Scopulariopsis candida Vuill., Bulletin de la Société Mycologique de France 27: 143 (1911) (Fig. 16).
≡Monilia candida Guég: 271 (1899).
≡Chrysosporium keratinophilum var. denticola C. Moreau, Mycopathologia et Mycologia Applicata 37 (1): 37 (1969).
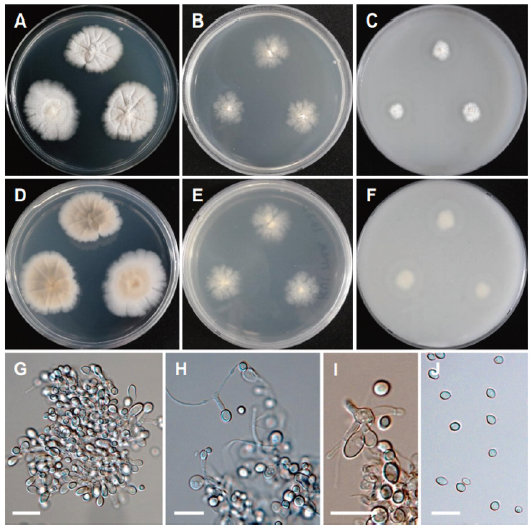
Fig. 16. Morphology of Scopulariopsis candida CNUFC KU1-1. (A, D) Colonies on potato dextrose agar (PDA). (B, E) Colonies on malt extract agar (MEA). (C, F) Colonies on oatmeal agar (OA). (A-C: Obverse view, D-F: Reverse view). (G-I) Conidiophores, annellides and conidia. (J) Conidia (scale bars=20 μm).
Description: Colonies on PDA irregular, white to cream-colored, reverse pale orange, measuring 24-28 mm after 7 d at 25℃. Colonies on MEA irregular, cream-colored, pale cream-colored, measuring 22-25 mm after 7 d at 25℃. Colonies on OA plane, regular, cottony white towards the center, reverse pale white, measuring 21-24 mm after 7 d at 25℃.
Micromorphology: Conidiophores were abundant. Conidia were subglobose to broadly ovate, hyaline smooth and measuring 4.3-8.3 × 4.4-7.3 μm.
Remarks: The genus Scopulariopsis is characterized by annellidic conidiogenesis with mostly thick-walled, basally truncate conidia arranged in long, dry chains and colony color ranging from white to brown or black [59]. Scopulariopsis candida is characterized by subglobose to broadly ovate, hyaline, smooth-walled conidia, irregularly ellipsoidal asci, broadly lunate to reniform ascospores and white colonies [35]. CNUFC KU1-1 differs from S. candida [35] by cream color colonies and no sexual morphology is observed. Previously S. candida has been reported from environmental samples (air, dust, and soil), predominantly in the Northern Hemisphere, especially Europe and North America; also isolated from clinical samples, mainly from superficial tissue of humans and animals suffering from onychomycosis [35,60,61]. This is the first report on the isolation of the species from seawater environment.
Volutella citrinella (Cooke &Massee) Seifert, Studies in Mycology 68: 110 (2011) (Fig. 17).
≡Stilbella aciculosa (Ellis & Everh.) Seifert, Studies in Mycology 37: 44 (1985).
≡Stilbum aciculosum Ellis & Everh., Journal of Mycology 1 (12): 153 (1885).
≡Botryonipha aciculosa (Ellis & Everh.) Kuntze, Revisio generum plantarum 2: 845 (1891).
≡Stilbella bulbicola Henn., Hedwigia 44: 176 (1905).
-Stilbum bulbicola (Henn.) M.A. Litv., Opredelitel' Mikroskopicheskikh Pochvennykh Gribov: 196 (1967).
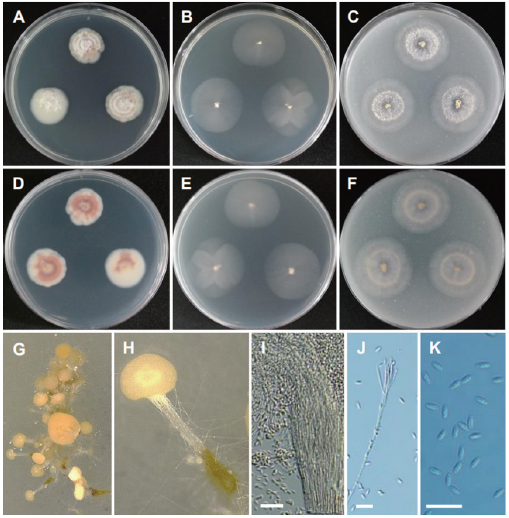
Fig. 17. Morphology of Volutella citrinella CNUFC DYR1-2. (A, D) Colonies on potato dextrose agar (PDA). (B, E) Colonies on malt extract agar (MEA). (C, F) Colonies on oatmeal agar (OA). (A-C: Obverse view, D-F: Reverse view). (G, H) Determinate synnemata developed in culture (observed under stereomicroscope). (I, J) Conidiophores. (K) Conidia (scale bars=20 μm).
-Stilbum bulbicola (Henn.) Sacc., Sylloge Fungorum 18: 633 (1906).
-Tubercularia bulbicola (Henn.) Sacc. (?).
≡Stilbella flavescens Estey, Transactions of the British Mycological Society 68 (1): 122 (1977).
≡Stilbum pallidulum Penz. & Sacc., Malpighia 15: 250 (1902).
≡Stilbum citrinellum Cooke & Massee, Grevillea 16 (79): 81 (1888).
-Volutella citrinella (Cooke & Massee) Seifert, Studies in Mycology 68: 110 (2011).
Description: Colonies on PDA floccose, zonate, white to creamy pink, reverse white to pink, measuring 21-25 mm after 7 d at 25℃. Colonies on MEA plane, regular, pale white, reverse pale white, measuring 25-26 mm after 7 d at 25℃. Colonies on OA plane, plane, regular, pale white to light creamy pink towards the center, reverse pale white, measuring 25-28 mm after 7 d at 25℃.
Micromorphology: Hyphae of synnema stipes thick-walled may appear seta-like when diverging from the synnema and measuring 1.3-3.0 μm wide with cell walls. Conidia were smooth and subglobose elliptical and measuring 4.1-9.3 × 2.2-2.6 μm.
Remarks: The genus Volutella includes species characterized by discoid sporodochia with marginal setae, compact and phialidic conidiogenous cells, simple to verticillate conidiophores, one-celled, ovoid to oblong conidia [19,62,63]. Volutella citrinella has been previously isolated from the debris of Solanum tuberosum and soil samples [9,41,64]. This is the first report on the occurrence of the species in a freshwater environment in Korea.
DISCUSSION
Phylogeny based on combined ITS-LSU rDNA and individual LSU rDNA sequences have placed seven isolates, CNUFC BCSM3, CNUFC F7-24-5, CNUFC HRG1, CNUFC HRW1-12, CNUFC YJS7, CNUFC PLTFB118, and CNUFC DYR1-2 with their respective type species of C. robusta, F. acetilerea, H. pedis, H. snookiorum, M. fusiformis, P. crystallina and V. citrinella, respectively. Since ITS do not aid in proper identification of genus Metarhizium and Scopulariopsis, TUB sequences for CNUFC AS1-26 and CNUFC KU1-1 were amplified [35,53,65]. Metarhizium pemphigi F1-72 (T) and FI-1101 isolated from root aphids lack the TUB gene [52], therefore Met. pemphigi isolates ARSEF 6569 and ARSEF 7491 of TUB gene have been used in the phylogeny [53,65]. Combined ITS-TUB phylogeny placed CNUFC KU1-1 with S. candida MUCL 40743 (T) [66].
Out of ten accepted Collariella species, only one species of Collariella carteri has been reported from Korea [67-70]. Species of Collariella were reported to produce a variety of metabolites, including chaetoquadrin E, cochliodinol B, cochliodones 1-3, prenisatin, and SB236049/SB236050/SB238569 [39,69,71]. Fusicolla comprises of 18 species, however only 14 of these are represented by rDNA sequences in GenBank [23,72-74]. Fusicolla merismoides was isolated from brackish water in Korea [75]. Fusicolla acetilerea was found to degrade triacrylonitrile (1,3,6-hexanetricarbonitrile) as a nitrogen source and 4-N-trimethylamino-1-butanol as a sole source of carbon and nitrogen [76,77]. There is no detailed study on the metabolites produced by four species described in Hongkongmyces [78]. According to the Index Fungorum 2021 (www.indexfungorum.org), the genus Mariannaea comprises of 23 species but only 18 species have GenBank records [49,79-81]. So far, three species have been reported from Korea [82]. Mariannaea species are known to produce extracellular enzymes such as amylases, β-glucosidases, proteases, and cellulases and thus can degrade cellobiase, starch and xylan [83,84]. Mariannaea elegans found to produce mariannamides A and B, new cyclic octapeptides and successfully mariannamides A showing antimicrobial activities against E. coli and C. neoformans [85]. Two species of Metarhizium have been reported from Korea from the accepted 34 species [86-88]. In particular, Metarhizium species have been exploited for the control of a wide range of arthropod pests such as plague locusts in Africa [89] as well as mosquito vectors of malaria [90]. Moreover Met. pemphigi is a promising biological control agent of ticks [91]. Eight Pallidocercospora species are accepted in this genus reported particularly as plant pathogens [92]. In total, 77 species are accepted in this genus [35,59] and 7 new species were introduced recently [69,93,94]. Scopulariopsis brevicaulis was found in Korea as a clinical isolate after cosmetic face surgery [95]. Scopulariopsis species known to cause deterioration of building materials [96,97] and accumulate various elements converting these into toxic volatiles [98,99] which makes them an important group to study in indoor environments. About 147 records of Volutella are listed in Index Fungorum 2021. with eight species having available sequences in GenBank [69,73,100]. Volutella ciliata was isolated from crop soil in Korea [101]. Volutella citrinella may be a candidate to produce new terpendole congeners N-P (1-3) and voluhemins that act as inhibitors of sterol-O-acyltransferase [64,102].
Results of this study clearly showed that there is relatively diverse group of species belonging to Dothideomycetes and Sordariomycetes associated with freshwater, soil, seawater, and plant samples from Korea. DNA sequences, especially those of the ITS and LSU rDNA regions have proven to be reliable for identifying species belonging to the Dothideomycetes and Sordariomycetes and some additional markers have also been introduced. The current study thus contributes to the knowledge on the fungal diversity from Dothideomycetes and Sordariomycetes in Korea with respect to their cosmopolitan distributions and environmental relevance. Future research is expected to cover the identification of important biochemical components and their activities and more genomes of Dothideomycetes and Sordariomycetes fungi become easily accessible and available.
ACKNOWLEDGMENTS
This study was financially supported by Chonnam National University [Grant number: 2017-2827]. This work was in part supported by the Graduate Program for the Undiscovered Taxa of Korea, the Project on Survey and Discovery of Indigenous Fungal Species of Korea funded by NIBR, and the Project on Discovery of Fungi from Freshwater and Collection of Fungarium, funded by NNIBR of the Ministry of Environment.