Introduction
Endophytic fungi do not elicit disease symptoms in their host plants when a balance is maintained among the environment, fungi, and plants [1]. Endophytic fungi are mainly belong to phylum Ascomycota [2] and form mutualistic relationships with host plants, thus providing them with benefits such as tolerance to environmental stress, pathogens, and secondary metabolites [3-5]. Endophytic fungi can be found in all plant tissues [6], and the structure of their communities varies depending on the parts and age of the tissues [7, 8]. Different fungal species inhabit different host plant tissues [9]. Foliar endophytes provide benefits such as antibacterial activity and defense against insect feeding on their host plants [10,11].
Approximately 55 conifer species have been recorded in Korea [12], which account for 42.4% of the total forest area [13]. Global warming has resulted in a worldwide decline in the conifer population [14,15]. The benefits conferred by endophytic fungi, including resistance to adverse environment, have the potential to improve the adaptability of conifers [16-18]. Therefore, it is important to investigate the diversity of endophytic fungi in conifers to conserve the forests. However, only a few studies have addressed this topic in Korea [19]. In the present study, the morphological and molecular characteristics of three species of previously unrecorded endophytic fungal isolates obtained from the leaves of two conifer species in Korea were investigated.
Materials and Methods
Samples from the two species of conifers—Pinus thunbergii Parl., and Chamaecyparis obtusa Siebold & Zucc.—were collected from Baengnyeongdo Island in Ongjin-gun, Incheon, and Palgeumdo Island in Sinan-gun, Jeollanam-do, respectively. Needle leaves without any symptoms of disease were selected and cut from their branches. The collected samples were transported to the laboratory in polyethylene bags within 24 h. First, the leaves were surface-sterilized with 1% sodium hypochlorite (NaOCl) for 1 min, followed by 70% ethanol (EtOH) for 2 min. The leaves were then cut into 1-2 cm pieces using sterile scissors and inoculated on potato dextrose agar (PDA) medium. Hyphae extending from the leaves were sub-cultured several times to obtain pure strains. The pure strains were identified based on their morphological and molecular characteristics. For morphological characterization, the fungal strains were cultured on malt extract agar (MEA) and PDA media in the dark at 25℃ for 7 days. Morphological characteristics such as texture, shape, and color of the colony and microstructures such as conidia and conidiophores were recorded. The sequences of the internal transcribed spacer (ITS) region, large subunit (LSU) region and elongation factor 1-alpha (TEF) gene were used for molecular characterization. Polymerase chain reaction (PCR) was performed using ITS1F/ITS4 [20], LROR/LR16 [21] and EF1-688F/EF1-1251R [22] primers. Amplicons of ITS, LSU and TEF were analyzed by SolGent Co. (Daejeon, Korea), and sequences were deposited in GenBank (National Center for Biotechnology Information). Phylogenetic trees were obtained using the combined ITS and LSU sequences with maximum likelihood phylogenetic analysis, which was conducted using MEGA 11 [23] with the Kimura-2 parameter distance model.
Results and Discussion
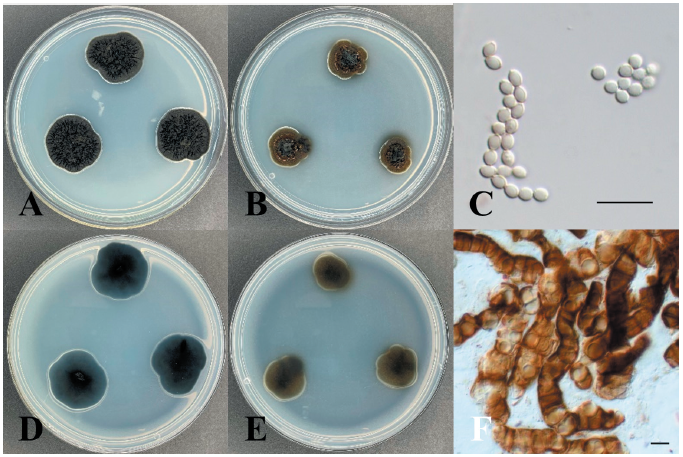
Celosporium laricicola Tsuneda & M.L. Davey, Botany 88: 473 (2010) [MB#548321] (Fig. 1)
Morphological characteristics of KNUE19E084: Morphological characteristics were observed after 7 days of incubation in the dark at 25℃. The diameter of the colonies cultured on the PDA medium was 15-17 mm, and the colonies showed black regular margins. The front side of the colonies was olive-yellow or dark olive-green and the reverse side was black in color. The colonies had a moist surface and exhibited furrows at the center. The color of the medium around the colonies changed to orange. The diameter of the colonies cultured on MEA medium was 8-11 mm and the colonies showed brown regular margins. Blastic conidia and endoconidia were lined together. Blastic conidia were ovoid, with a size of (2.21-) 2.78 (-3.23)×(1.69-) 1.98 (-2.36) µm. Endoconidia consisted of 1-3 globular cells, and their size was (6.90-) 10.36 (-12.35) × (5.89-) 7.83 (-10.36) µm. For morphological analyses, 20 conidia were identified, and MEA medium was used for conidia production.
Specimen examined: Baengnyeongdo Island, Ongjin-gun, Incheon, Republic of Korea; 37°57'N, 124°38'E, May 15, 2019, isolated from the leaves of Pinus thunbergii, NIBR No. NIBRFGC000508554, GenBank No. ITS region - ON430568, LSU region - ON430567.
Note: Celosporium laricicola was isolated from twigs of Larix lyallii by Tsuneda & Davey in 2010 [24]. Celosporium comprises only one species, Celosporium laricicola. It is characterized by the formation of endoconidia composed of 1–3 cells within a cellular clump. The ITS region of KNUE19E084 showed 99.01% similarity with that of FJ997287, and the LSU region showed 99.66% similarity with that of FJ997288. Through morphological and molecular analysis, KNUE19E084 discovered in this study, was identified as Celosporium laricicola, an unrecorded species in Korea (Fig. 2).
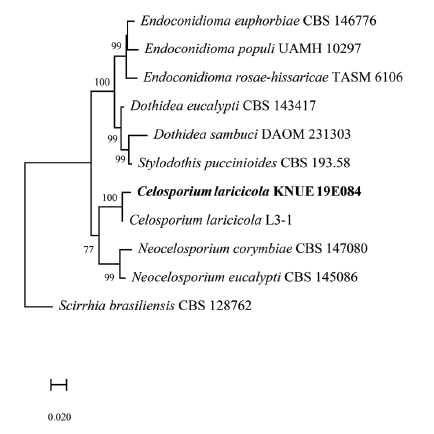
Fig. 2.Maximum likelihood phylogenetic analysis of combined internal transcribed spacer (ITS)and large subunit (LSU) of rDNA sequences of Celosporium laricicola KNUE19E084. Scirrhia brasiliensis was used as an outgroup. Numbers on branches indicate bootstrap values (1,000 replicates). Fungal strain isolated in this study is in a bold.
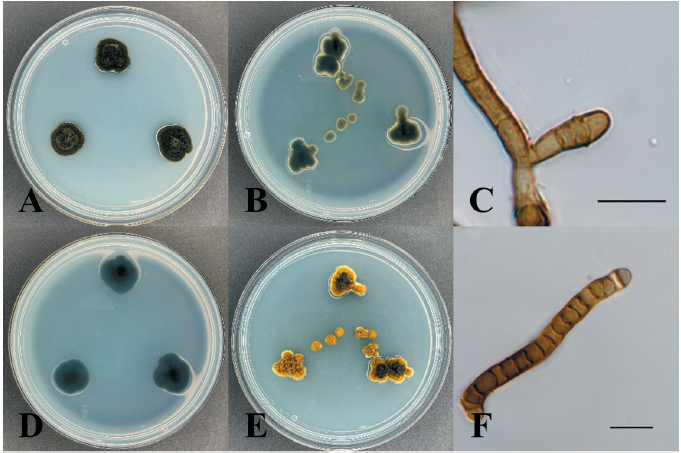
Neocatenulostroma germanicum (Crous & U. Braun) Quaedvl. & Crous, Persoonia 33: 27 (2014) [MB#807807] (Fig. 3)
Morphological characteristics of KNUE19E267: Morphological characteristics were observed after 7 days of incubation in the dark at 25℃. The diameters of the colonies on the PDA and MEA media were 8-10 and 7-8 mm, respectively. The colonies showed regular margins. The front side of the colonies had a deep olive-green color, and the reverse side was black on PDA medium. Colonies on the MEA medium appeared slightly brighter than those on the PDA medium. Furrows appeared in the colonies, with dents and cracks around the colonies. The color of the conidia was dark brown, and their size was (16.74-) 19.22 (-24.40)×(4.42-) 5.34 (-6.58) µm. They had septa, and their features were simple or branched basipetal chain, straight or flexuous, smooth, and sub-cylindrical. For morphological analysis, 20 conidia were identified, and MEA medium was used for conidia production.
Specimen examined: Baengnyeongdo Island, Ongjin-gun, Incheon, Republic of Korea; 37°57'N, 124°38'E, May 15, 2019, isolated from the leaves of Pinus thunbergii; NIBR No. NIBRFGC000508555, GenBank No. ITS region - ON430577, LSU region - ON430565.
Notes: In 2007, this species was first recorded by Crous et al. [25], isolated from stone, and named Catenulostroma germanicum. Markovskaja et al. [26] isolated it as a pathogen from Pinus spp. and reclassified it as Neocatenulostroma germanicum. The sequences of the ITS region of KNUE19E267 showed 99.41% similarity with those of MK622897, and the LSU region showed 99.66% sequence similarity with that of MH873835. The TEF gene, which was analyzed for a more accurate identification showed 97.8% similarity with KU612279. Through morphological and molecular analyses, KNUE19E0267, discovered in this study, was identified as Neocatenulostroma germanicum, an unrecorded species in Korea (Fig. 4).
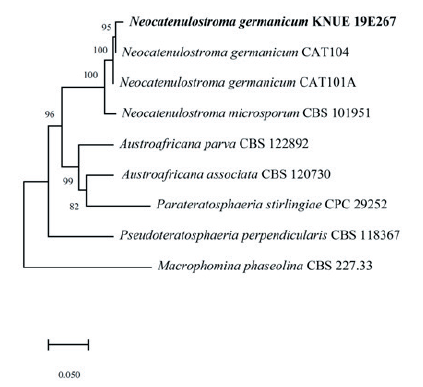
Fig. 4.Maximum likelihood phylogenetic analysis of combined internal transcribed spacer (ITS), large subunit (LSU) of rDNA and elongation factor 1-alpha (TEF) sequences of Neocatenulostroma germanicum KNUE19E267. Macrophomina phaseolina was used as an outgroup. Numbers on branches indicate bootstrap values (1,000 replicates). Fungal strain isolated in this study is in a bold.
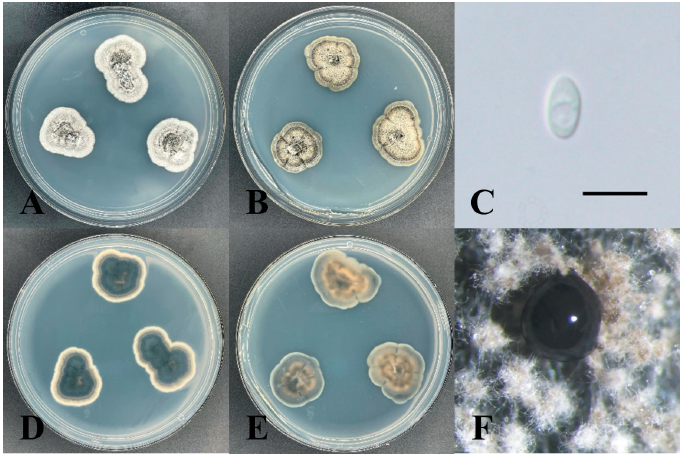
Phaeophleospora eucalypticola Crous & M.J. Wingf., Persoonia 36: 351 (2016) [MB#817030] (Fig. 5)
Morphological characteristics of KNUE19E083: Morphological characteristics were observed after 7 days of incubation in the dark at 25℃. The diameters of the colonies on both PDA and MEA media were 14 mm. The colonies exhibited furrows and irregular margins. The color of the colonies on the PDA medium was light gray on both the front and back sides and yellowish-gray on MEA medium. The size of the conidia was (2.21-) 2.78 (-3.23)×(1.69-) 1.98 (-2.36) µm. The conidia were hyaline, and morphological features such as a smooth surface, ellipsoid, and base truncate were observed. For morphological analyses, 20 conidia were identified, and MEA medium was used for conidia production.
Specimen examined: Palgeumdo Island, Sinan-gun, Jeollanam-do, Republic of Korea; 34°44'N, 126°7'E, April 11, 2019, isolated from the leaves of Chamaecyparis obtusa NIBR No. NIBRFGC000505588, GenBank No. ITS region - ON430538, LSU region - ON430542.
Note: Phaeophleospora eucalypticola was first reported by Crous et al. [27] and was isolated from the leaves of Eucalyptus grandis × Eucalyptus camaldulensis clone. The sequences of the ITS region of KNUE19E083 showed 99.64% similarity with those of NR_145123, and the sequence of the LSU region showed 99.67% similarity with those of NG_069357. The TEF gene, analyzed for a more accurate identification showed 98.03% similarity with KX228374. Through morphological and molecular analyses, KNUE19E083 discovered in this study, was identified as Phaeophleospora eucalypticola, an unrecorded species in Korea (Fig. 6).
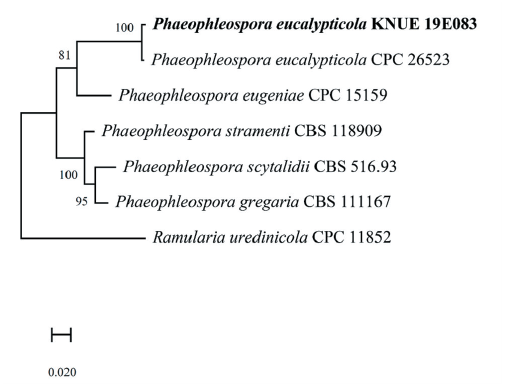
Fig. 6.Maximum likelihood phylogenetic analysis of combined internal transcribed spacer (ITS), large subunit (LSU) of rDNA and elongation factor 1-alpha (TEF) sequences of Phaeophleospora eucalypticola KNUE19E083. Ramularia uredinicola was used as an outgroup. Numbers on branches indicate bootstrap values (1,000 replicates). Fungal strain isolated in this study is in a bold.