Puccinia is the largest genus of the family Pucciniaceae (Pucciniomycetes), which comprises approximately 4,000 species [1], all of which are obligate plant pathogens and cause the rust diseases on ecologically and economically important crops and trees. Recently, Puccinia klugkistiana has been recorded on an ornamental shrub, Ligustrum japonicum, in Korea [2]. Previously, it was known that P. klugkistiana occurs in spermogonial and aecial stages on L. obtusifolium, and in uredinial and telial stages on Cleistogenes hackelii [3]. However, so far there is no record of the occurrence of this rust on C. hackelii in Korea. The prerequisite for promoting an effective control measure against P. klugkistiana is to find its potential alternative host in Korea. Through our field foray, we found hundreds of C. hackelii (Honda) Honda (Poaceae), which was associated with a rust fungus in October 2019. Some leaves were dominated by uredinia, and others by telia, allowing the microscopic examinations and molecular analysis. To the best of our knowledge, this is the first study to report the occurrence of both uredinial and telial stages of rust on C. hackelii in Korea. Three specimens have been deposited in the Korea University Herbarium (KUS) with the accession numbers of KUS-F30372 (9 XI 2017, Daegu, Korea; stage III), F31285 (15 X 2019, ibid.; stage II), and F31286 (15 X 2019, ibid.; stage III). To elucidate the identity of the rust, morphological examination and molecular phylogenetic analyses were performed.
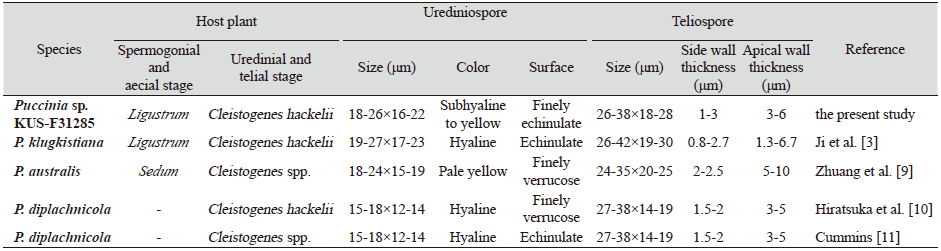
The detailed morphological characteristics of this rust were observed using an Olympus BX50 microscope (Olympus, Tokyo, Japan). The surface structures of the rust were observed using a field-emission scanning electron microscope (SUPRA 55VP, Carl Zeiss, Germany). Uredinia were hypophyllous, scattered, orange-yellow in color, and more or less erumpent through the host cuticle. Urediniospores were subglobose to ellipsoid in shape, subhyaline to yellowish in color, finely echinulate, and 18-26×16-22 μm in size. Telia were hypophyllous, distinctly erumpent through the host cuticle, dark brown to blackish, and devoid of paraphyses. Teliospores were subglobose, two-celled, more or less constricted at the septa, 26-38×18-28 μm in size, with 46- 88 μm long pedicel, and wall thicknesses were 1-3 μm on the sides and 3-6 μm at the apices These characteristics were concordant with those of P. klugkistiana [3].
To confirm the morphological identity of this rust, genomic DNA was extracted from the teliospores of KUS-F31285 (Korea University Herbarium, Seoul, Korea) using the DNeasy Plant Mini Kit (Qiagen, Hilden, Germany) according to the manufacturer’s instructions. PCR was performed to amplify the internal transcribed spacer (ITS) region and large subunit (LSU) rDNA using the specific primer pair ITS5u/ITS4rust and universal primers LR0R/LR6, respectively [4]. The PCR products were purified using the AccuPrep® PCR Purification Kit (Bioneer, Daejeon, Korea) and sequenced through the DNA sequencing service (Macrogen, Seoul, Korea) with the same primers used for the amplification. The resulting sequences were edited using the Lasergene software package version 5.05 (DNASTAR, Madison, WI, USA) and registered to the GenBank database (accession no. MW740211 for ITS and MW740212 for LSU). BLASTn search revealed that they are identical to P. klugkistiana ex L. japonicum (MH644179 and MH644180 for ITS; MH644181 and MH644182 for LSU). ITS and LSU sequences were aligned with the reference sequences of Puccinia species in NCBI GenBank using MAFFT 7 [5]. To construct the phylogenetic trees, maximum likelihood (ML) analysis was performed using MEGA 7 [6], through applying the Tamura-Nei model with 1,000 bootstrap replicates. In the phylogenetic trees for ITS (Fig. 2A) and LSU (Fig. 2B), the present rust fungus was grouped with the sequences of P. klugkistiana occuring on L. japonicum with high supporting values of 99 and 100%, thereby confirming its identity as P. klugkistiana.
Till date, four different species of Puccinia have been recorded on Cleistogenes hackelii, viz., P. klugkistiana, P. australis, P. diplachnicola, and P. permixta [3,7]. Previously, Hiratsuka et al. [8] recorded P. diplachnicola on C. hackelii based on the two collections (Sep 10 and Oct 3, 1941, Y. Fujikuro, Seoul, Korea). However, the spores of P. diplachnicola are much smaller than those of P. klugkistiana, e.g., 15-18×12-14 μm versus 18-26×16-22 μm for urediniospores and 38×14-19 μm versus 26-38×18-26 μm for teliospores (Table 1). Moreover, aecial hosts of P. diplachnicola have been reported as species of Sedum in Japan [12] and China [9]. Although there has been no previous record of Sedum sp. as an aecial host of P. diplachnicola in Korea, we still conclude that the rust species isolated from C. hackelii in this study is unrelated to P. diplachnicola. This is the first report of P. klugkistiana on C. hackelii in Korea.

Fig. 1. Puccinia klugkistiana on Cleistogenes hackelii. A: Rust symptoms on the leaf surface. B: Uredinia on the leaf surface. C: Telia on the leaf surface. D: Coexistence of uredinia (U) and telia (T). E: Erumpent telia. F: Urediniospores (u), immature (it) and mature (mt) teliospores observed under a light microscope. G and H: A telium with a group of teliospores (G) and close-up of teliospores (H) observed under a scanning electron microscope.
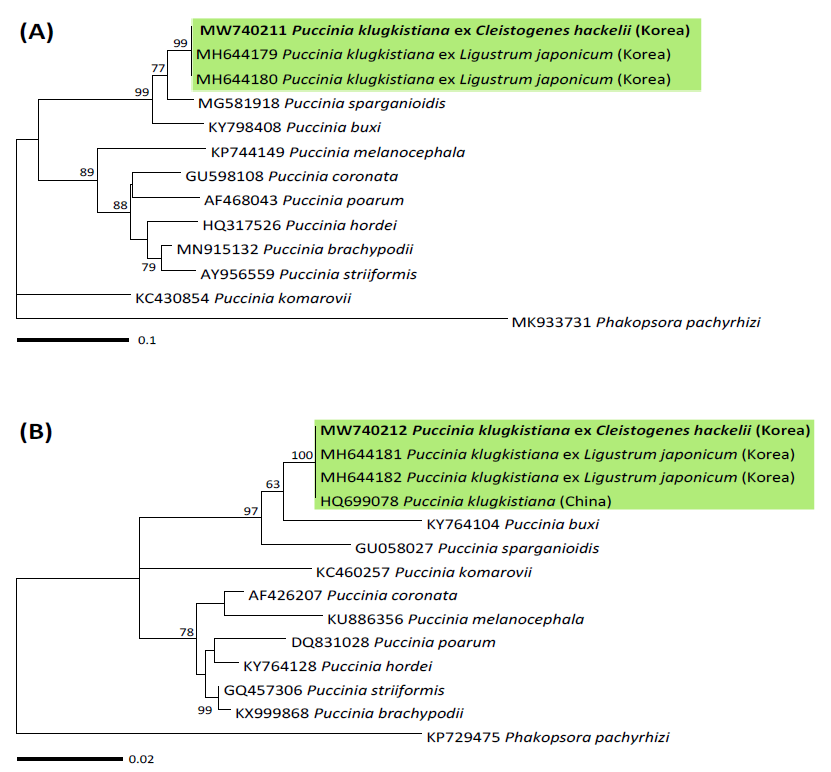
Fig. 2. Maximum likelihood phylogenetic tree of Puccinia species based on the internal transcribed spacer (ITS) (A) and large subunit (LSU) (B) rDNA sequences. The robustness was evaluated using 1,000 bootstrap replicates, and the numbers above the branches of the phylogenetic tree represent the bootstrap values over 50%. The Korean specimen sequenced in the present study is indicated in bold.