Introduction
Hypocreales is one of the largest orders within the class Sordariomycetes in Ascomycota. More than 2,647 species (seven families and 237 genera) belong to this order [1]. Fungal species in this order generally show brightly colored perithecial ascomata or spore-producing structures. They are found worldwide and in various habitats, including aquatic and terrestrial environments. As described in the National List of Species of Korea, 212 species in this order have been reported in Korea until 2020 [2].
The genus Dactylonectria belongs to the family Nectriaceae, one of the largest families of Hypocreales, Ascomycota. To date, 18 species belonging to this genus have been reported [3]. Several species of this genus have been reported as plant pathogens and endophytic fungi [4]. This genus is classified in terms of its anamorphic stage and conidial morphology [5].
In the genus Ilyonectria of the family Nectriaceae, 37 species have been reported in 2021 [3]. Various species of this genus have been reported as saprophytes on decaying wood [4]. This genus is classified according to the characteristics of its sexual stage and conidia morphology [5-7].
In the genus Nectria of the family Nectriaceae, 938 species have been reported in 2021 [3]. Several species of this genus have been reported as plant soilborne pathogens [4]. This genus is classified by characteristics such as sexual stage and mycological morphology, such as color and texture [8-10].
The genus Neonectria belongs to the family Nectriaceae. To date, 52 species belonging to this genus have been reported [3]. Several species of this genus are common in tropical and temperate regions and have been reported as woody plant pathogens [11]. Occasionally, they have been found in decaying materials [4,12]. This genus is classified according to characteristics such as sexual stage and conidial morphology [8].
In the genus Pseudonectria of the family Nectriaceae, 40 species have been reported to date [3]. Several species of this genus are well-known plant pathogens [13]. Occasionally, they have been found in decaying materials [4]. This genus is classified according to characteristics such as sexual stage and conidial morphology [4,14].
The genus Fusarium is one of the largest genera in the family Nectriaceae, and more than 1,000 species have been reported in 2021. Several species of this genus have been reported as plant pathogens in the air and soil [15]. This genus is classified according to the characteristics of sexual stage and morphology of asexual conidia [16].
The genus Scolecofusarium of the family Nectriaceae has been classified as a new combination in 2021. To date, only one species belonging to this genus has been reported [3]. This genus is classified according to its unique sexual stage and pionnotes on the agar surface [17].
The genus Flavocillium belongs to Cordycipitaceae, one of the largest families of Hypocreales, and only four species belonging to this genus have been reported [18]. All species of this genus were in the genus Lecanicillium until 2020 and have recently been introduced as a new genus by multigene phylogeny [18]. Some species of this genus have been reported as entomopathogenic fungi [18]. This genus is classified by its sexual and asexual stages [18].
In the genus Lecanicillium of the family Cordycipitaceae, more than 21 species have been reported [19]. Some species of this genus have been reported as entomopathogenic fungi [19] and have been used as commercial biological pesticides [20]. This genus is classified by an asexual stage [19].
The genus Ovicillium belongs to Bionectriaceae, one of the common families of Hypocreales, and was classified as a new combination in 2016 [21]. To date, only four species have been reported [3]. Fungal species of this genus are found in mushrooms, wood, plants, and soil [21]. This genus is classified according to characteristics such as asexual stage and conidial morphology [21].
The genus Sarocladium belongs to Sarocladiaceae, one of the families of Hypocreales, and was classified as a new combination in 2016 [21]. Thirty species have been reported to date [3]. Several species of Sarocladium have been reported as plant pathogens or as saprobes or human pathogens [22]. This genus is classified based on characteristics such as the asexual stage, conidial morphology, and molecular phylogeny [22].
The genus Trichoderma belongs to Hypocreaceae, one of the largest families of Hypocreales, and this genus is one of the largest genera in Hypocreales. To date, 483 species have been reported [3]. Several species of this genus are known to be potential biocontrol agents for plant pathogens, and studies on antimicrobial activities have been carried out for a long time [23]. This genus is classified based on the morphological characteristics of the asexual stage [24].
To the best of our knowledge, this is the first report of 17 Hypocreales species (Dactylonectria hordeicola, Flavocillium bifurcatum, Fusarium luffae, Ilyonectria ilicicola, Ilyonectria qitaiheensis, Ilyonectria robusta, Lecanicillium aphanocladii, Nectria ulmicola, Neonectria lugdunensis, Ovicillium oosporum, Pseudonectria foliicola, Sarocladium spinificis,, Scolecofusarium ciliatum, Trichoderma appalachiense, Trichoderma subviride, Trichoderma taiwanense, and Trichoderma tsugarense) in Korea from environmental samples such as freshwater, plant litter, sediment in rivers, wetland, streams, and forest soil; the molecular, phylogenetic and morphological characteristics of these species were also investigated.
Materials and Methods
Isolation of fungal strains and culture conditions
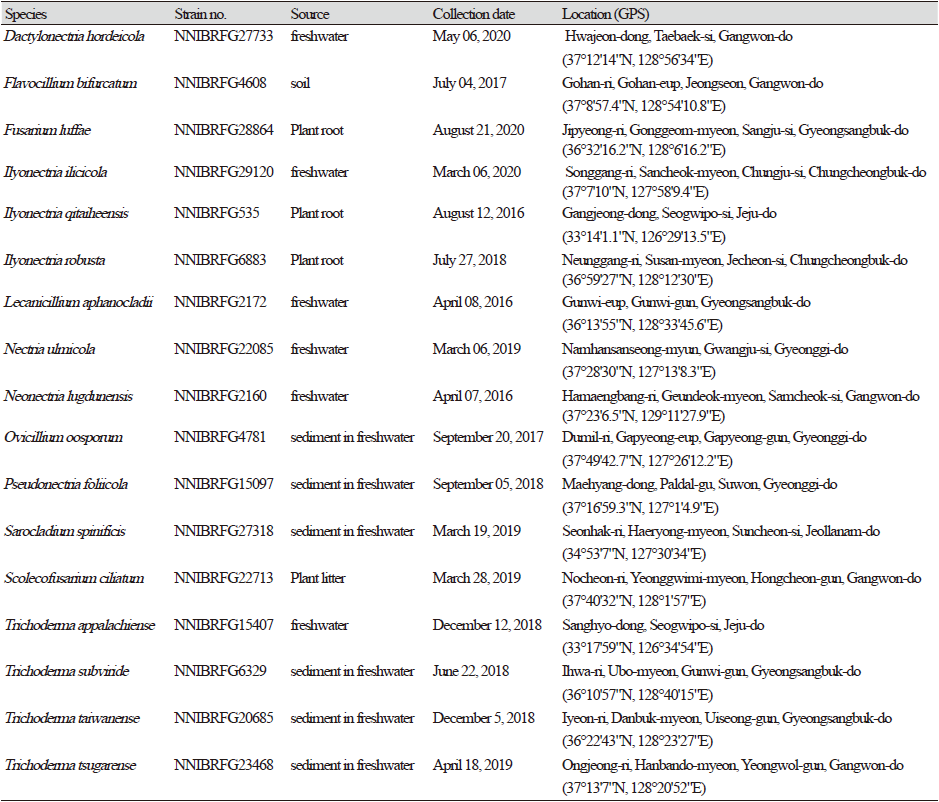
Fungal strains were collected from environmental samples, including freshwater and forests, in Korea. The collection information for all strains identified in this study is listed in Table 1. To isolate fungal strains, plant litter samples were washed with distilled water at least twice and incubated in a pretreatment liquid medium (0.05% 3-morpholinopropane-1-sulfonic acid [weight/volume (w/v)], 0.05% KNO3 [w/v], 0.025% KH2PO4 [w/v], and 0.025% K2HPO4 [w/v]) at 20℃ for 3 d. Thereafter, 100 µL of the pretreatment medium was spread on a 1% water agar plate and incubated at 20℃ for 2 d. Hyphal tips and germinated conidia were isolated under a microscope, transferred onto a 24-well plate containing V8 agar (V8A; 8% V8 juice [v/v] and 1.5% agar [w/v], adjusted to pH 6.0, using 10 N NaOH), and incubated at 25℃ in the dark. A dilution method was used to isolate fungal strains from sediment and soil. The diluted suspensions (200 μL) of sediment or soil with distilled water (1:200 and 1:2,000) were spread on potato dextrose agar (PDA; 3.9% PDA powder [w/v]; Difco, Sparks, MD, USA) containing 50 ppm streptomycin, and fungal strains were isolated in pure form after incubation for 4-5 d at 25℃ by repeating this step. All strains identified in this study were grown on malt extract agar (MEA; 2% malt extract [w/v] and 2% agar [w/v]), oatmeal agar (OA; 7.25% OA powder [w/v]; Difco), synthetic nitrogen-poor or nutrient-poor agar (SNA; 0.02% sucrose [w/v], 0.02% glucose [w/v], 0.1% KNO3 [w/v], 0.1% KH2PO4 [w/v], 0.05% MgSO4·7H2O [w/v], 0.05% NaCl [w/v], and 1.2% agar [w/v]), corn meal dextrose agar (CMDA; 2% cornmeal [w/v], 2% glucose [w/v], and 2% agar), and yeast extract peptone dextrose agar (YPDA; Duchefa Biochemie, Haarlem, Netherland).
DNA extraction, polymerase chain reaction (PCR), and DNA sequencing
Fungal genomic DNA was isolated using the NucleoSpin® Plant II DNA extraction kit (Macherey-Nagal, Düren, Germany). For molecular identification of fungi, PCR amplification was performed for the internal transcribed spacer (ITS) rDNA region using primers ITS1 (5′-TCCGTAGGTGAACCTGCGG-3′) and ITS4 (5′-TCCTCCGCTTATTGATATGC-3′) [25], for the large subunit of rDNA (LSU) using primers LR0R (5′-ACCCGCTGAACTTAAGC-3’) and LR7 (5′-TACTACCACCAAGATCT-3′) [26], for the beta-tubulin gene (TUB) using primers bt2a (5′-GGTAACCAAATCGGTGCTGCTTTC-3′) and bt2b (5′-ACCCTCAGTGTAGTGACCCTTGGC-3′) [27], for the translation elongation factor 1 gene (EF1) using primers EF1-983F (5′-GCYCCYGGHCAYCGTGAYTTYAT-3′) and EF1-1576R (5′-ACHGTRCCRATACCACCRATCTT-3′) [28], and for the RNA polymerase II gene (RPB2) using primers fRPB2-5F (5′-GAYGAYMGWGATCAYTTYGG-3′) and fRPB2-7cR (5′-CCCATRGCTTGYTTRCCCAT-3′) [29]. Amplicons were sequenced with the help of a DNA sequencing service (Macrogen Inc., Seoul, Korea) using the same primers as those used for amplification. A homology search of DNA sequences was performed using BLAST algorithms available from the National Center for Biotechnology Information (NCBI; https://www.ncbi.nlm.nih.gov).
Phylogenetic analysis
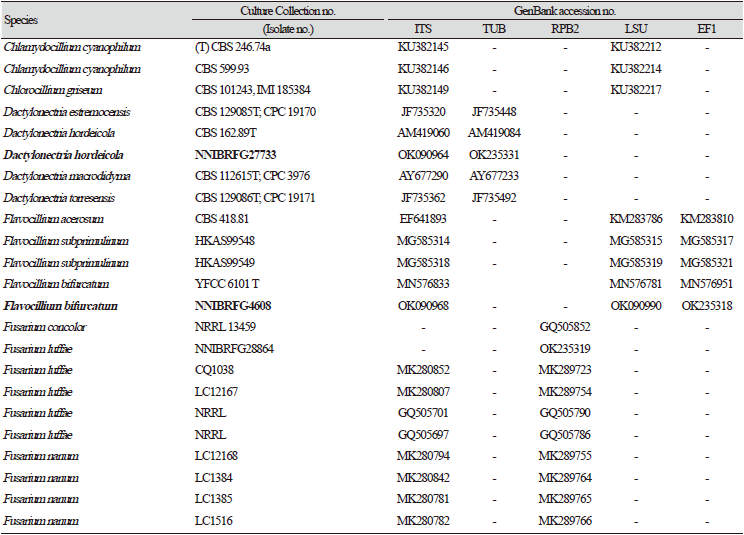
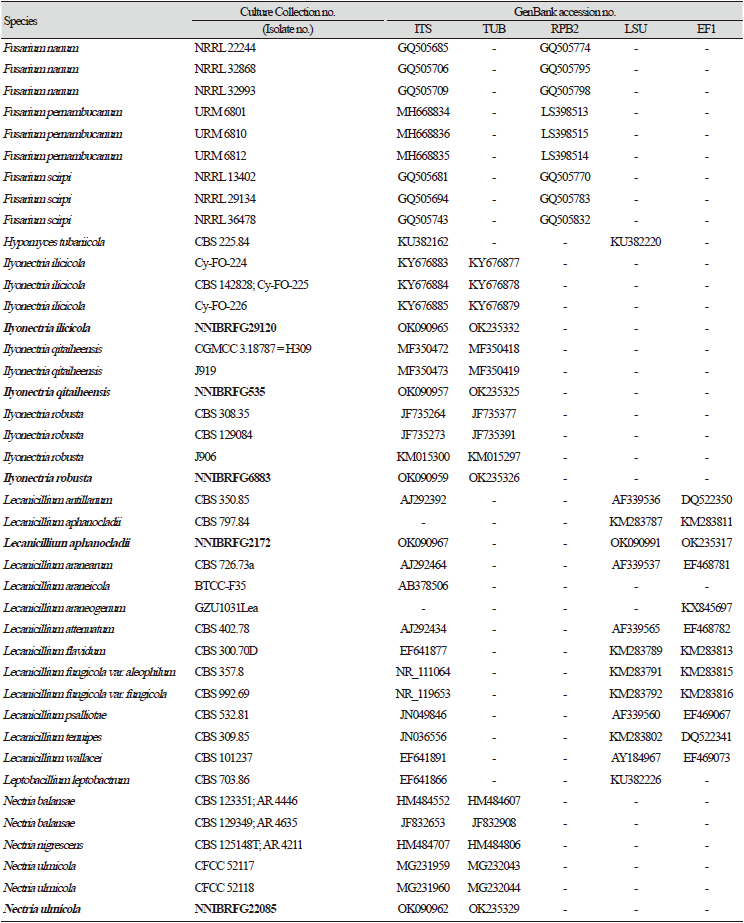
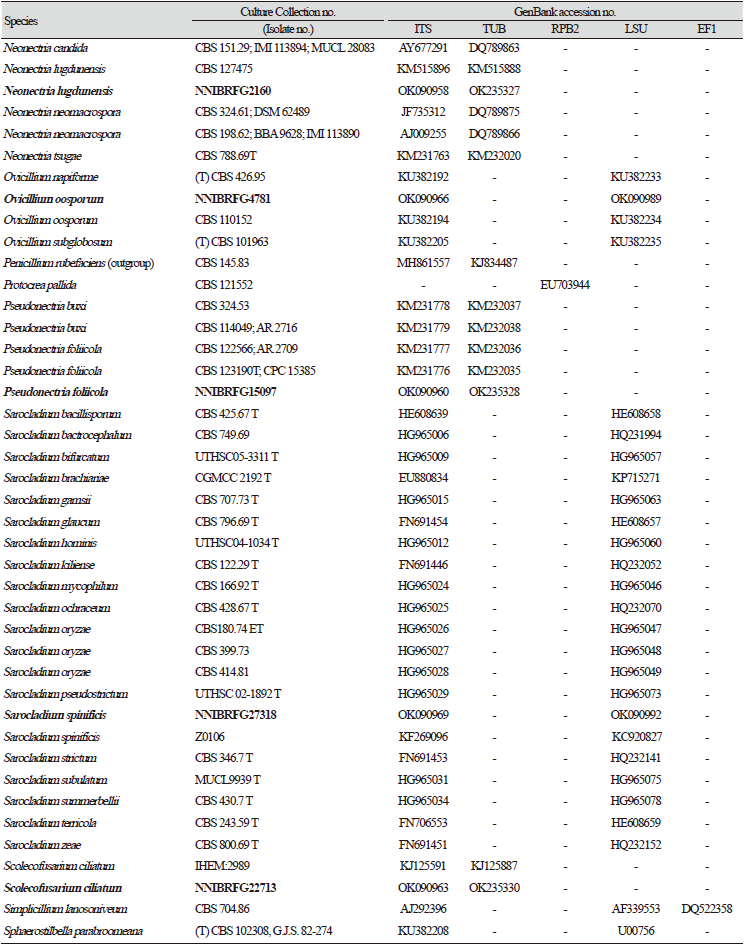
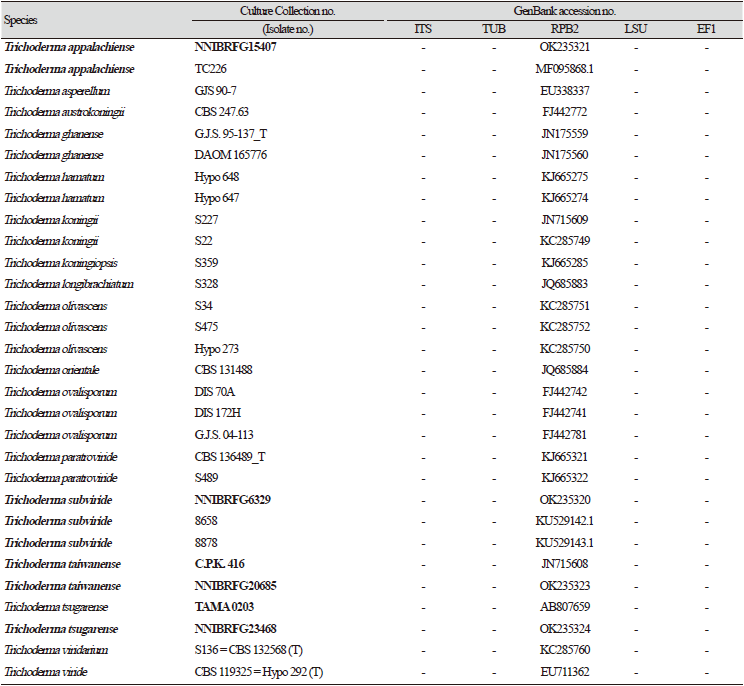
Previously published reference sequences [4-24,30-45] were obtained from the NCBI (Table 2). Sequences were edited using the DNAStar software package version 5.05 (DNAStar, Inc., Madison, WI, USA). The accession numbers of sequences used in this study are shown in phylogenetic trees (Figs. 1-6) and Table 2. Phylogenetic trees were constructed using the maximum likelihood (ML) anlalysis, which was performed using MEGA 7.1 [46] with the default settings of the program, except for replacement with the Tamura-Nei model. Bootstrapping analysis of 1,000 replicates was performed to test the robustness of each grouping.
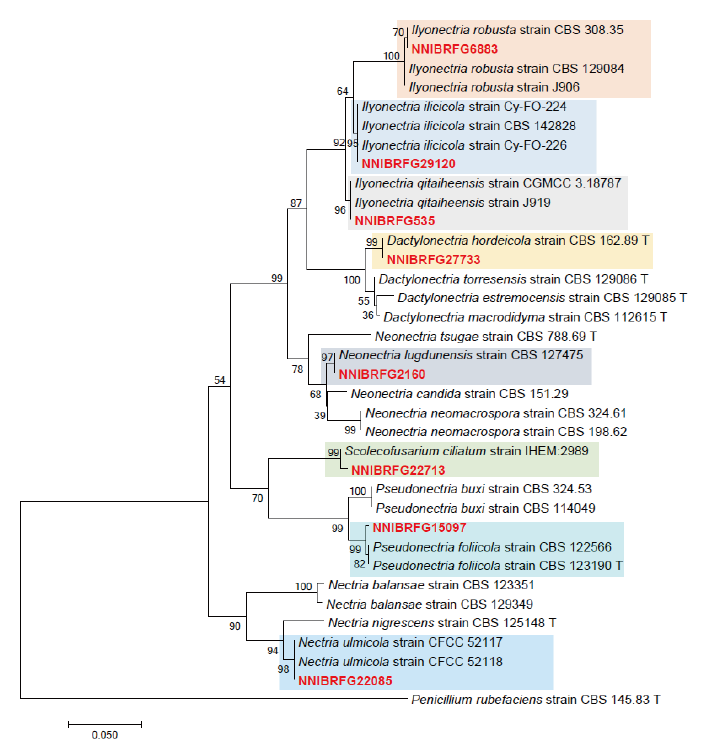
Fig. 1. Phylogenetic tree of Dactylonectria, Ilyonectria, Nectria, Neonectria, Pseudonectria, Scolecofusarium and their related species, based on maximum-likelihood analysis of the combination of internal transcribed region sequences and beta-tubulin gene sequences. Numbers at the nodes indicate the bootstrap values (>50%) from 1,000 replications. Red color font indicates fungal strains in this study.
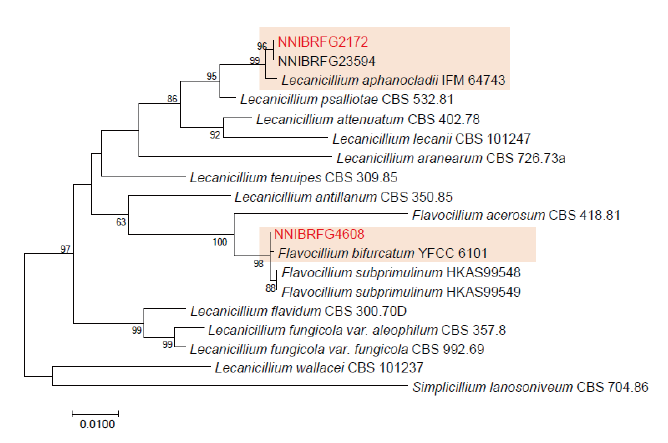
Fig. 3. Phylogenetic tree of Lecanicillium, Flavocillium and their related species, based on maximum-likelihood analysis of the combination of internal transcribed region sequences, the large subunit of rDNA sequences and elongation factor 1 sequence. Numbers at the nodes indicate the bootstrap values (>50%) from 1,000 replications. Red color font indicates fungal strains in this study.
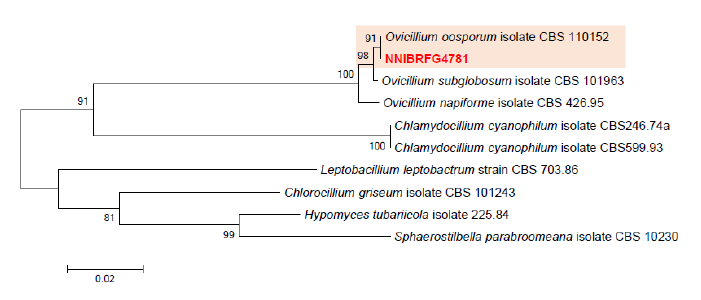
Fig. 4. Phylogenetic tree of Ovicillium and its related species, based on maximum-likelihood analysis of the combination of internal transcribed region sequences and the large subunit of rDNA sequences. Numbers at the nodes indicate the bootstrap values (>50%) from 1,000 replications. Red color font indicates fungal strain in this study.
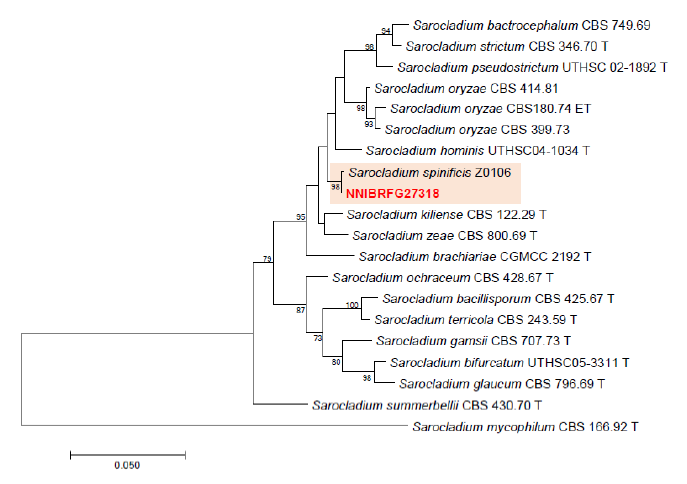
Fig. 5. Phylogenetic tree of Sarocladium and its related species, based on maximum-likelihood analysis of the combination of internal transcribed region sequences and the large subunit of rDNA sequences. Numbers at the nodes indicate the bootstrap values (>50%) from 1,000 replications Red color font indicates fungal strain in this study.
Morphological analysis
Microstructures of fungal species were observed under an Eclipse Ni light microscope (Nikon, Tokyo, Japan) equipped with a Ds-Ri2 digital camera (Nikon). At least 50 individuals were examined for the observation and measurement of each structure. For scanning electron microscopy, we followed the protocol previously described by Alves et al. [47]. Photographs were obtained using a scanning electron microscope (SEM; SU8220, Hitachi, Japan).〮〮〮〮
Analysis of antifungal, enzyme, and plant growth-promoting activities
For the antifungal activity assay, we used two plant pathogens, Fusarium oxysporum and F. solani, from Angelica gigas in Youngju (Forest Medicinal Resources Research Center). Inhibition rates were measured using a dual culture assay at 7 dpi at 25℃. To determine chitinase activities, discoloration was observed 7 d after inoculation on media with the following supplements: 4.5 g colloidal chitin, 0.3 g MgSO4 · 7H2O, 3 g NH4SO4, 1 g KH2PO4, 0.15 g bromocresol purple, 0.2 mL Tween 80, and 15 g/L agar [48]. To determine laccase activity, discoloration was observed 7 d after inoculation on media supplemented with 5 g malt extract, 0.2 g guaiacol, and 15 g/L agar [49]. To determine other enzyme activities, halo zones were measured on minimal MEA (1% malt extract [w/v] and 1.5% agar [w/v]) supplemented with the appropriate substrate after 2 weeks of incubation at 25℃. The supplemented substrates included 0.5% Tween 20 (w/v) for lipase activity and 1% skim milk (w/v) for protease activity in the presence of Congo red (500 ppm). For the phosphate solubilization assay, a clear zone was observed 7 d after inoculation on Pikovskaya agar (HiMedia, Mumbai, India) [50]. For the siderophore production assay, discoloration was observed 7 d after inoculation on CAS media [51].
Results and Discussion
Phylogenetic analysis
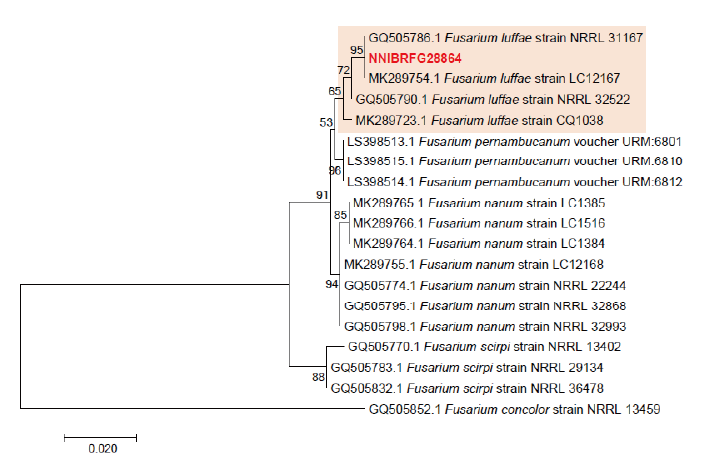
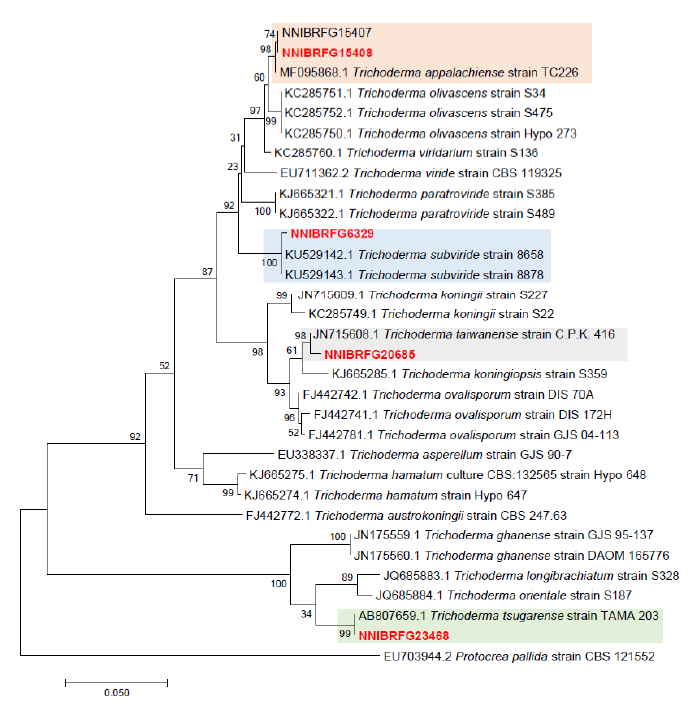
Phylogenetic analysis was performed to identify the 17 fungal strains and infer their phylogenetic relationships with other similar species. ITS and TUB sequences were used for phylogenetic analysis of Dactylonectria, Ilyonectria, Nectria, Neonectria, Pseudonectria, Scolecofusarium, and their related species. As shown in Fig. 1, strain NNIBRFG6883 formed a clade with three other strains of Ilyonectria robusta. The sequences of NNIBRFG6883 showed high similarities (100% in ITS and 99.69% in TUB) with those of I. robusta. NNIBRFG29120 formed a clade with three other strains of I. ilicicola. The sequences of NNIBRFG29120 showed high similarity (100% in ITS and 99.63% in TUB) with those of I. ilicicola. NNIBRFG535 formed a clade with two other strains of I. qitaiheensis. The sequences of the NNIBRFG535 showed high similarities (100% in ITS and 99.69% in TUB) with those of I. qitaiheensis. NNIBRFG27733 formed a clade with a strain of Dactylonectria hordeicola. The sequences of NNIBRFG27733 showed high similarities (100% in ITS and TUB) with those of D. hordeicola. NNIBRFG2160 formed a clade with the Neonectria lugdunensis strain. The sequences of the NNIBRFG2160 showed high similarities (100% in ITS and TUB) with those of N. lugdunensis. NNIBRFG22713 formed a clade with the S. ciliatum strain. The sequences of NNIBRFG22713 showed high similarities (99.39% in ITS and 99.37% in TUB) with those of S. ciliatum. NNIBRFG15097 formed a clade with two other strains, including Pseudonectria foliicola. The sequences of NNIBRFG15097 showed high similarities (100% in ITS and 98.16% in TUB) with those of Pseudonectria foliicola. NNIBRFG22085 formed a clade with two other strains of Nectria ulmicola. The sequences of NNIBRFG22085 showed high similarities (100% in ITS and TUB) with those of Nectria ulmicola. As shown in Fig. 2, RPB2 sequences were used for the phylogenetic analysis of Fusarium species. NNIBRFG28864 formed a clade with four strains of Fusarium luffae. The sequences of NNIBRFG28864 showed a high similarity (100% in RPB2) with those of F. luffae. As shown in Fig. 3, a combination of ITS, LSU, and EF1 sequences was used for the phylogenetic analysis of Lecanicillium, Flavocillium, and their related species. NNIBRFG4608 formed a clade with Flavocillium bifurcatum. BLASTn analysis of NNIBRFG4608 genes showed high similarities (99.8% [ITS], 99.87% [LSU], and 99.57% [EF1]) with those of F. bifurcatum. NNIBRFG2172 formed a clade with a strain of L. aphanocladii. BLASTn analysis of NNIBRFG2172 genes showed high similarities (100% [ITS], 100% [LSU], and 98.87% [EF1]) with those of L. aphanocladii. As shown in Fig. 4, a combination of ITS and LSU sequences was used for the phylogenetic analysis of Ovicillium and the related species. NNIBRFG4781 formed a clade with a strain of Ovicillium oosporum. The sequences of NNIBRFG4781 showed high similarities (100% in ITS and 99.64% in LSU) with those of O. oosporum. As shown in Fig. 5, a combination of ITS and LSU sequences was used for the phylogenetic analysis of Sarocladium and its related species. NNIBRFG27318 formed a clade with a strain of Sarocladium spinificis,. The sequences of NNIBRFG27318 showed high similarities (100% in ITS and 99.80% in LSU) with those of S. spinificis. As shown in Fig. 6, RPB2 sequences were used for the phylogenetic analysis of Trichoderma species and the related species. NNIBRFG15408 formed a clade with a T. appalachiense strain. The sequences of NNIBRFG15408 showed a high similarity (99.89% in RPB2) with those of T. appalachiense. NNIBRFG6329 formed a clade with two strains of T. subviride. The sequences of NNIBRFG6239 showed a high similarity (99.77% in RPB2) with those of T. subviride. NNIBRFG20685 formed a clade with a T. taiwanense strain. The sequences of NNIBRFG20685 showed a high similarity (99.48% in RPB2) with those of T. taiwanense. NNIBRFG23468 formed a clade with a T. tsugarense strain. The sequences of NNIBRFG23468 showed a high similarity (100% in RPB2) with those of T. tsugarense.
Through the molecular identification, we identified 17 fungal species in Hypocreales. Moreover, we examined antifungal activities for plant pathogens, extracellular enzyme activities, and plant growth promotion activities (Table 3). These results suggested that many species of this study could be potential biocontrol agents or other industrial usage.
In this study, here we report the characterization of unrecorded fungal species from environmental samples. It could contribute to understanding fungal diversity and characterization of various habitat in Korea.
Table 3. Antifungal activities, enzyme activities, and plant growth promotion of the strains used in this study.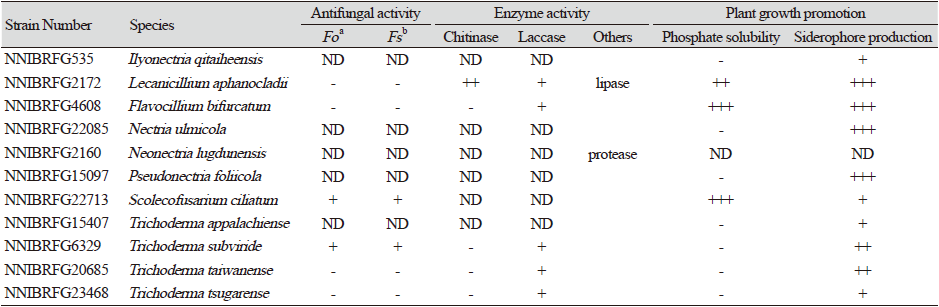
|
Taxonomy
Dactylonectria hordeicola L. Lombard & Crous, Phytopathologia Mediterranea 53 (3): 527 (2014) [MB#810146] (Figs. 7A and 8A-D)
Description: Colonies grew slowly at 25 ℃ and reached 11 mm on CMDA, 11 mm on MEA, 19 mm on OA, 10 mm on PDA, 23 mm on V8A, and 19 mm on YPDA, 7 d after inoculation. The color of the colony was hyaline with a smooth aerial mycelial surface on CMDA, hyaline with a smooth aerial mycelial surface on MEA, creamy white with fluffy aerial mycelia on OA, creamy white to light yellow from the center with fluffy aerial mycelia on PDA, creamy white to brown with cottony aerial mycelia on V8A, and creamy white to yellow with fluffy aerial mycelia on YPDA. Microconidia were aseptate or 1 septate, hyaline, cylindrical, measuring 8.9-20.9 µm×2.9-5.8 μm (x=13.6±2.57 µm×4.1±0.62 μm, L/W ratio=3.32, n=50). Chlamydospores were brownish and globose or subglobose, measuring 7.7-15.0×7.2-13.7 μm (x=11.2±2.76 µm×10.1±2.53 μm, n=10).
Habitat: Filtered freshwater in a stream
Specimen examined: Hwajeon-dong, Taebaek-si, Gangwon-do, 06 May, 2020, NNIBRFG27733, Nakdonggang National Institute of Biological Resources
Note: Dactylonectria hordeicola was first reported as an isolate from Hordeum vulgare [4]. In this study, NNIBRFG27733 was isolated from filtered freshwater streams.
Flavocillium bifurcatum H. Yu, Y.B. Wang, Y. Wang, Q. Fan & Zhu L. Yang, Fungal Diversity 103: 20 (2020) [MB#833096] (Figs. 9A and 10A-B)
Description: Colonies grew well at 25℃ and reached 27 mm on CMD, 27 mm on MEA, 29 mm on OA, 26 mm on PDA, 31 mm on V8A, and 16 mm on MEA, 7 d after inoculation. The color of the colony was hyaline with a cottony aerial mycelial surface on CMDA, hyaline with a cottony aerial mycelial surface on MEA, creamy white with dense aerial mycelia on OA, creamy white with dense aerial mycelia on PDA, creamy white with fluffy aerial mycelia on V8A, and creamy white with dense aerial mycelia on YPDA. Following 10 d of growth on PDA, a diffusing pink-red pigment was observed. Conidia were hyaline, aseptate, and ovoid to ellipsoid, measuring 4.0-17.1 µm×1.8-4.0 μm (x=7.7±2.84 µm×2.7±0.51 μm, L/W ratio=2.86, n=50).
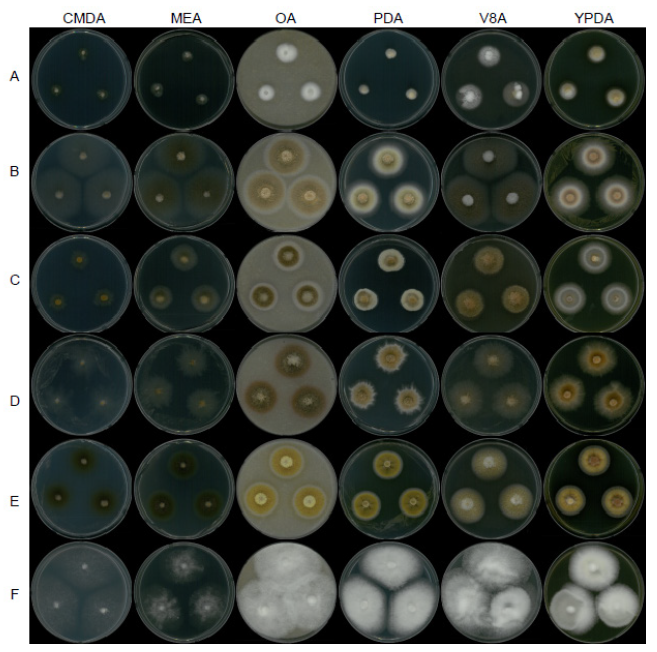
Fig. 7. Mycelial growth on corn meal dextrose agar (CMDA), malt extract agar (MEA), oatmeal agar (OA), potato dextrose agar (PDA), V8 agar (V8A), and yeast extract peptone dextrose agar (YPDA) for 7 days at 25℃. A, Dactylonectria hordeicola strain NNIBRFG27733; B, Ilyonectria ilicicola strain NNIBRFG29120; C, Ilyonectria qitaiheensis strain NNIBRFG535; D, Ilyonectria robusta strain NNIBRFG6883; E, Neonectria lugdunensis strain NNIBRFG2160; F, Pseudonectria foliicola strain NNIBRFG15097.
Habitat: Soil in forest
Specimen examined: Gohan-ri, Gohan-eup, Jeongseon, Gangwon-do, 04 July 2017, NNIBRFG4608, Nakdonggang National Institute of Biological Resources
Note: Flavocillium bifurcatum was isolated from larva of Noctuidae [18]. In this study, NNIBRFG4608 was isolated from soil in forests. Notably, NNIBRFG4608 showed enhanced siderophore production, phosphate solubility, and laccase activity (Table 3).
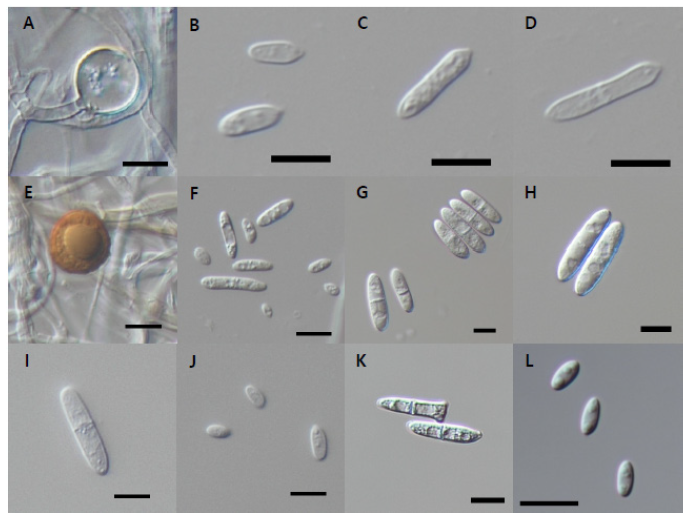
Fig. 8. Microscopic observation. A-D, Conidial morphology of Dactylonectria hordeicola strain NNIBRFG27733. Chlamydospore (A) and conidia (B-D); E-F, Ilyonectria ilicicola strain NNIBRFG29120. E, chlamydospore (E) and conidia (F); G-H, conidia morphology of Ilyonectria qitaiheensis strain NNIBRFG535; I-J, conidia morphology of Ilyonectria robusta strain NNIBRFG6883; K, conidia morphology of Neonectria lugdunensis strain NNIBRFG2160; L, conidia morphology of Pseudonectria foliicola strain NNIBRFG15097. (Scale bars: 10 μm).
Fusarium luffae M.M. Wang, Qian Chen & L. Cai, Persoonia 43: 85 (2019) [MB#829540] (Figs. 9C and 10E-F)
Description: The colonies grown at 25℃ reached 16 mm on CMDA, 28 mm on MEA, 59 mm on OA, 41 mm on PDA, 73 mm on V8A, and 72 mm on YPDA, 7 d after inoculation. The color of the colony was hyaline with a few aerial mycelia on the surface of CMDA, hyaline to white from the center with a smooth aerial mycelial surface on MEA, light yellow with fluffy aerial mycelia on PDA, creamy white to light yellow from the center with fluffy aerial mycelia on PDA, hyaline and creamy white with cottony aerial mycelia on V8A, and creamy white with abundant aerial mycelia on YPDA. Macroconidia were 2 to 7 septate, hyaline, falcate, straight to curved, and measured 20.6-41.5 µm×3.9-6.1 μm (x=32.5±4.76 µm×4.9±0.52 μm, L/W ratio=6.62, n=50). Chlamydospores were not observed in this study.
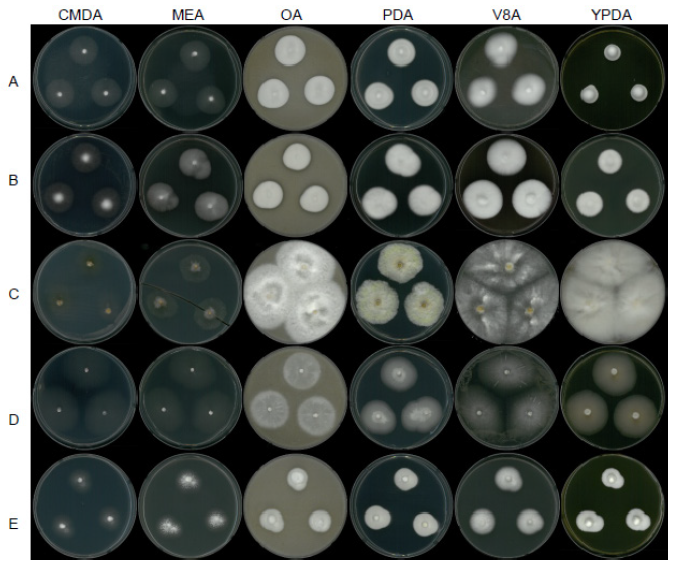
Fig. 9. Mycelial growth on corn meal dextrose agar (CMDA), malt extract agar (MEA), oatmeal agar (OA), potato dextrose agar (PDA), V8 agar (V8A), and yeast extract peptone dextrose agar (YPDA) for 7 days at 25℃. A, Flavocillium bifurcatum strain NNIBRFG4608; B, Lecanicillium aphanocladii strain NNIBRFG2172; C, Fusarium luffae strain NNIBRFG28864; D, Ovicillium oosporum strain NNIBRFG4781; E, Sarocladium spinificis, strain NNIBRFG27318.
Habitat: Root of Viola verecunda in a stream
Specimen examined: Jipyeong-ri, Gonggeom-myeon, Sangju-si, Gyeongsangbuk-do, 21 August 2021, NNIBRFG28864, Nakdonggang National Institute of Biological Resources
Note: Fusarium luffae was initially isolated from Luffa aegyptiaca [16], but in this study, three strains were isolated from the roots of Viola verecunda.
Ilyonectria ilicicola B. Mora-Sala, A. Cabral, Armengol & Abad-Campos, Plant Disease 102 (11): 2095 (2018) [MB#822025] (Figs. 7B and 8E-F)
Description: Colonies grew slightly fast at 25℃ and reached 50 mm on CMDA, 48 mm on MEA, 43 mm on OA, 37 mm on PDA, 50 mm on V8A, and 39 mm on YPDA, 7 d after inoculation. The color of the colony was hyaline with a cottony aerial mycelial surface on CMDA, hyaline to brown with a cottony aerial mycelial surface on MEA, white to dark goldenrod with fluffy aerial mycelia on OA, white to goldenrod from the center with fluffy aerial mycelia on PDA, grey-brown with cottony aerial mycelia on V8A, and creamy white to dark goldenrod with fluffy aerial mycelia on YPDA. Macroconidia were 1-3 septate, hyaline, cylindrical, and measured 18.0-32.6 µm×3.9-5.8 μm (x=23.0±4.48 µm×4.38±1.03 μm, L/W ratio=5.2, n=50). Microconidia were aseptate or 1 septate, hyaline, fusiform, and measured 3.9-7.7 µm×2.3-3.6 μm (x=5.9±1.05 µm×3.0±0.33 μm, L/W ratio=1.93, n=50). Chlamydospores were brownish and globose or subglobose, measuring 13.8-20.6 μm×12.9-19.4 μm (x=17.4±1.77 µm×16.2±1.70 μm, n=50).
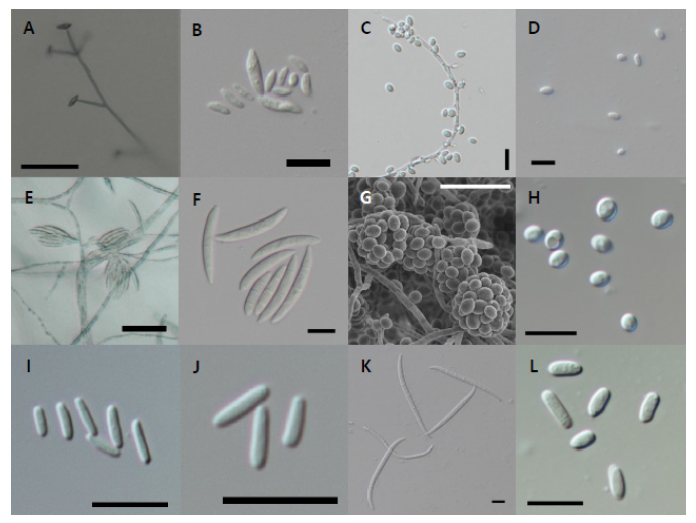
Fig. 10. Microscopic observation. A-B, Conidiophore (A) and conidia (B) morphology of Flavocillium bifurcatum strain NNIBRFG4608; C-D, conidiophore (C) and conidia (D) morphology of Lecanicillium aphanocladii strain NNIBRFG2172; E-F, conidiophore (E) and conidia (F) morphology of Fusarium luffae strain NNIBRFG28864; G-H, conidiophore (G) and conidia (H) morphology of Ovicillium oosporum strain NNIBRFG4781; I-J, conidia morphology of Sarocladium spinificis, strain NNIBRFG27318; K, conidia morphology of Scolecofusarium cilliatum strain NNIBRFG22713; L, conidia morphology of Nectria ulmicola strain NNIBRFG22085. Scale bars are 10 μm (B-D, F-L) and 50 μm (A and E).
Habitat: Filtered freshwater from a stream
Specimen examined: Songgang-ri, Sancheok-myeon, Chungju-si, Chungcheongbuk-do, Republic of Korea, 6 March 2020, NNIBRFG29120, Nakdonggang National Institute of Biological Resources
Note: Ilyonectria ilicola was isolated from the roots of Ilex sp. [6]. In this study, three strains were isolated from filtered freshwater from streams.
Ilyonectria qitaiheensis X. Lu & W. Gao, Journal of Ginseng Research 44: 514 (2020) [MB#823895] (Figs. 7C and 8G-H)
Description: Colonies grew slightly slow at 25℃ and reached 16 mm on CMDA, 43 mm on MEA, 29 mm on OA, 30 mm on PDA, 19 mm on V8A, and 18 mm on YPDA at 25℃, 7 days after inoculation. The color of the colony was hyaline with a smooth aerial mycelial surface on CMDA, hyaline to grey-brown with cottony aerial mycelial surface on MEA, white to dark goldenrod from center with fluffy aerial mycelia on OA; creamy white to goldenrod from center with fluffy aerial mycelia on PDA, yellow brown with fluffy aerial mycelia on V8A, and creamy white to grey-brown with cottony aerial mycelia on YPDA. Macroconidia were 1 to 3 septate, hyaline, cylindrical, and measured 19.5-35.3 µm×4.6-8.3 μm (x=28.7±3.72 µm×6.8±0.74 μm, L/W ratio=4.17, n=50).
Habitat: Root of aquatic plant (Umbelliterae sp.) near a stream
Specimen examined: Gangjeong-dong, Seogwipo-si, Jeju-do, Republic of Korea, March 6, 2020, NNIBRFG535, Nakdonggang National Institute of Biological Resources
Note: Ilyonectria qitaiheensis was isolated from the roots of Panax ginseng in China [4]. In this study, NNIBRFG535 was isolated from the roots of aquatic plants (Umbelliterae sp.) near the stream.
Ilyonectria robusta (A.A. Hildebr.) A. Cabral & Crous, Mycological Progress 11 (3): 680 (2011) [MB#560113] (Figs. 7D and 8I-J)
Description: Colonies grew slightly slow at 25℃ and reached 17 mm on CMDA, 30 mm on MEA, 36 mm on OA, 30 mm on PDA, 36 mm on V8A, and 36 mm on YPDA, 7 d after inoculation. The color of the colony was hyaline with rough margins on CMDA, hyaline to grey-brown with a cottony aerial mycelial surface on MEA, dark goldenrod with fluffy aerial mycelia on OA, brown from the center with an ivory rough margin on PDA, hyaline to light grey with cottony aerial mycelia on V8A, and light brown to dark goldenrod with dense aerial mycelia on YPDA. Macroconidia were 1-3 septate, hyaline, cylindrical with round ends, and measured 19.5-35.3 µm×4.6-8.3 μm (x=23.9±6.47 µm×6.4±1.17 μm, L/W ratio=3.97, n=50). The 1-septate macroconidia measured 9.6-32.0×2.7-8.3 μm (x=22.9±5.71 µm×5.9±1.16 μm, L/W ratio=3.87). The 2-septate macroconidia measured 25.9-38.1×5.2-8.5 μm (x=30.9±4.13 µm×6.8±1.13 μm, L/W ratio=4.52). The 3-septate macroconidia measured 30.7-38.6×5.6-8.9 μm (x=34.7±3.07 µm×7.1±0.88 μm, L/W ratio=4.89). Microconidia were aseptate or 1 septate, hyaline, ellipsoid, measuring 4.4-12.0 µm×2.2-4.8 μm (x=8.0±1.61 µm×3.5±0.59 μm, L/W ratio=2.25, n=50). Chlamydospores were brownish and globose or subglobose, measuring 10.8-17.6 μm×9.0-15.7 μm (x=13.7±1.69 µm×12.5±1.67 μm, n=50).
Habitat: Root of moss on rock near a stream
Specimen examined: Neunggang-ri, Susan-myeon, Jecheon-si, Chungcheongbuk-do, Republic of Korea, 7 July 2018, NNIBRFG6883, Nakdonggang National Institute of Biological Resources
Note: Ilyonectria robusta was isolated from the living root of Panax quinquefolium in Canada [7]. In this study, NNIRFG6883 was isolated from the root of moss in a rock near a stream.
Lecanicillium aphanocladii Zare & W. Gams, Nova Hedwigia 73 (1-2): 27 (2001) [MB#484541] (Figs. 9B and 10C-D)
Description: Colonies grew well at 25℃ and reached 28 mm on CMD, 26 mm on MEA, 27 mm on OA, 30 mm on PDA, 36 mm on V8A, and 24 mm on MEA, 7 d after inoculation. The color of the colony was hyaline to creamy white with a fluffy aerial mycelial surface on CMDA, white with a cottony aerial mycelial surface on MEA, creamy white with dense aerial mycelia on OA, creamy white with dense aerial mycelia on PDA, creamy white with dense aerial mycelia on V8A, and creamy white to light pink from the margin with dense aerial mycelia on YPDA. After a 10 d incubation on PDA, a diffusing pink-red pigment was observed. Conidia were produced solitary at the tip of aphanophialides, were aseptate, oval to subglobose, and measured 3.0-8.5 µm×1.5-4.2 μm (x=5.3±1.26 µm×2.7±0.60 μm, L/W ratio=1.95, n=50).
Habitat: Freshwater from a stream
Specimen examined: Gunwi-eup, Gunwi-gun, Gyeongsangbuk-do, 08 April 2016, NNIBRFG2172, Nakdonggang National Institute of Biological Resources
Note: Several strains of L. aphanocladii were isolated from mushrooms and leaf litter [19]. In this study, NNIBRFG2172 was isolated from freshwater stream. Notably, NNIBRFG2172 showed enhanced siderophore production and phosphate solubility, as well as other extracellular enzyme activities, such as chitinase, laccase, and lipase (Table 3).
Nectria ulmicola C.M. Tian & Q. Yang, Phytotaxa 356 (3): 204 (2018) [MB#825504] (Figs. 10Land 11B)
Description: Colonies grew slowly at 25℃ and reached 22 mm on MEA, 18 mm on OA, and 12 mm on PDA, 7 d after inoculation. The color of the colony was hyaline with a few aerial mycelia on the surface of MEA, hyaline to white with short cottony aerial mycelia on OA, creamy white with a rough margin, and short cottony aerial mycelia on PDA. Conidia were hyaline, cylindrical or ellipsoid, aseptate, and measured 4.8-8.7 µm×2.3-3.6 μm (x=6.4±0.89 µm×3.0±0.32 μm, L/W ratio=2.16, n=50).
Habitat: Filtered freshwater from a stream
Specimen examined: Namhansanseong-myun, Gwangju-si, Gyeonggi-do, 06 Mar 2019, NNIBRFG22085, Nakdonggang National Institute of Biological Resources
Note: Nectria ulmicola was isolated from twigs and branches of Ulmus davidiana var. japonica in China [10]. In this study, NNIBRFG22085 was isolated from filtered freshwater from a stream. Notably, NNIBRFG22085 showed enhanced siderophore production (Table 3).
Neonectria lugdunensis (Sacc. & Therry) L. Lombard & Crous, Phytopathologia Mediterranea 53 (3): 528 (2014) [MB#810155] (Figs. 7E and 8K)
Description: Colonies grew well at 25℃ and reached 25 mm on CMD, 28 mm on MEA, 34 mm on OA, 28 mm on PDA, 34 mm on V8A, and 29 mm on MEA, 7 d after inoculation. The color of the colony was dark olive green with a cottony aerial mycelial surface on CMDA, dark olive green with a cottony aerial mycelial surface on MEA, yellow to light yellow from the center with fluffy aerial mycelia on OA, goldenrod to brown with dense aerial mycelia on PDA, light brown with fluffy aerial mycelia on V8A, and yellow orange to goldenrod from the center with dense aerial mycelia on YPDA. Macroconidia were hyaline, 0–3 septate, clavate with a widened subapical region, and measured 17.5-30.4 µm×3.7-6.4 μm (x=24.8±3.07 µm×5.2±0.76 μm, n=50).
Habitat: Filtered freshwater from a stream
Specimen examined: Hamaengbang-ri, Geundeok-myeon, Samcheok-si, Gangwon-do, 07 April 2016, NNIBRFG2160, Nakdonggang National Institute of Biological Resources
Note: Neonectria lugdunensis was first isolated from submerged decaying alder leaves in a stream bed of England [12]. In this study, NNIBRFG2160 was isolated from filtered freshwater from streams. Moreover, NNIBRFG2160 showed weak protease activity (Table 3).
Ovicillium oosporum Zare & W. Gams, Mycological Progress 15: 1022 (2016) [MB#815498] (Figs. 9D and 10G-H)
Description: Colonies grew slightly fast at 25℃ and reached 43 mm on CMDA, 46 mm on MEA, 36 mm on OA, 35 mm on PDA, 51 mm on V8A, and 41 mm on MEA, 7 d after inoculation. The color of the colony was hyaline with a short cottony aerial mycelial surface on CMDA, hyaline with a cottony aerial mycelial surface on MEA, creamy white with fluffy aerial mycelia on OA, hyaline to creamy white from the center, with fluffy aerial mycelia on PDA, hyaline to white with cottony aerial mycelia on V8A, and white to light lemon with smooth aerial mycelia on YPDA. Conidia were hyaline, aseptate, and subglobose to globose, measuring 3.6-5.5 µm×2.6-4.5 μm (x=4.2±0.47 µm×3.6±0.46 μm, L/W ratio=1.18, n=50).
Habitat: Sediment in freshwater
Specimen examined: Dumil-ri, Gapyeong-eup, Gapyeong-gun, Gyeonggi-do, Republic of Korea, 20 September 2017, NNIBRFG4781, Nakdonggang National Institute of Biological Resources
Note: Ovicillium oosporum was isolated from Theobroma gileri in South America; however other strains were also identified, as this species had various isolation sources such as soil, humans, mushrooms, insects, etc., [21]. In this study, NNIBRFG4781 was isolated from sediment in streams.
Pseudonectria foliicola L. Lombard & Crous, Studies in Mycology 80: 219 (2015) [MB#810180] (Figs. 7F and 8L)
Description: Colonies grew fast at 25℃ and reached 51 mm on CMDA, 37 mm on MEA, 61 mm on OA, 61 mm on PDA, 63 mm on V8A, and 45 mm on MEA, 7 d after inoculation. The color of the colony was hyaline to white with a cottony aerial mycelial surface on CMDA, hyaline to white with a cottony aerial mycelial surface and a rough margin on MEA, creamy white with abundant fluffy aerial mycelia on OA, creamy white with fluffy dense aerial mycelia on PDA, creamy white with fluffy aerial mycelia on V8A, and creamy white with fluffy dense aerial mycelia on YPDA. Conidia were hyaline, aseptate, and fusiform to ellipsoid, measuring 4.7-8.2 µm×2.1-3.6 μm (x=6.4±0.81 µm×2.9±0.33 μm, L/W ratio=2.22, n=50).
Habitat: sediment in freshwater
Specimen examined: Dumil-ri, Gapyeong-eup, Gapyeong-gun, Gyeonggi-do, Republic of Korea, 20 September 2017, NNIBRFG4781, Nakdonggang National Institute of Biological Resources
Note: Pseudonectria foliicola was isolated from the leaves of Buxus sempervirens in New Zealand [14] and reported as a pathogen of boxwood causing Volutella blight [13]. In this study, NNIBRFG15097 was isolated from sediment in streams. Notably, NNIBRFG15097 showed enhanced siderophore production (Table 3).
Sarocladium spinificis, Yu Hung Yeh & R. Kirschner, Botanical Studies 55 (25): 3 (2014) [MB#805250] (Figs. 9E and 10I-J)
Description: Colonies grew slightly slow at 25℃ and reached 22 mm on CMDA, 24 mm on MEA, 24 mm on OA, 23 mm on PDA, 23 mm on V8A, and 23 mm on MEA, 7 d after inoculation. The color of the colony was hyaline to white from the center with a cottony aerial mycelial surface on CMDA, creamy white with a cottony aerial mycelial surface on MEA, creamy white with dense aerial mycelia on OA, creamy white with dense aerial mycelia on PDA, white with fluffy aerial mycelia on V8A, and creamy white with dense aerial mycelia on YPDA. Conidia were hyaline, aseptate, cylindrical or ellipsoid, and measured 3.9-8.3 µm×1.2-3.0 μm (x=5.7±0.88 µm×1.8±0.32 μm, L/W ratio=3.19, n=50).
Habitat: Sediment in freshwater
Specimen examined: Seonhak-ri, Haeryong-myeon, Suncheon-si, Jeollanam-do, Republic of Korea, 19 March 2019, NNIBRFG27318, Nakdonggang National Institute of Biological Resources
Note: Sarocladium spinificis, was isolated as an endophytic fungus from the coastal grass Spinifex littoreus in Taiwan [40]. In this study, NNIBRFG2731s8 was isolated from sediment in streams.
Scolecofusarium ciliatum (Link) L. Lombard, Sand.-Den. & Crous, Studies in Mycology 98 (no. 100116): 74 (2021) [MB#838677] (Figs. 10K and 11A)
Description: Colonies grew slowly at 25℃ and reached 18 mm on PDA, 20 mm on OA, and 21 mm on MEA, 7 d after inoculation. The color of the colony was light yellow with a smooth aerial mycelial surface on PDA, hyaline with a smooth aerial mycelial surface on MEA, and apricot white to light goldenrod from the center with slightly fluffy aerial mycelia on OA. Macroconidia were slightly falcate, smooth-walled, and 3-7 septate, measuring 41.9-92.7 µm×3.1-5.3 μm (x=68.6±9.9 µm×3.9±0.4 μm, L/W ratio=17.47, n=50).
Habitat: Plant litter in stream
Specimen examined: Nocheon-ri, Yeonggwimi-myeon, Hongcheon-gun, Gangwon-do, 28 March 2019, NNIBRFG22713, Nakdonggang National Institute of Biological Resources
Note: Previous molecular phylogenetic studies have suggested that the position of S. ciliatum can be moved to another genus [34,36]. Several strains of S. ciliatum have been reported as isolates from the leaves of Fagus sylvatica [17]. In this study, NNIBRFG22713 was isolated from plant litter in streams.
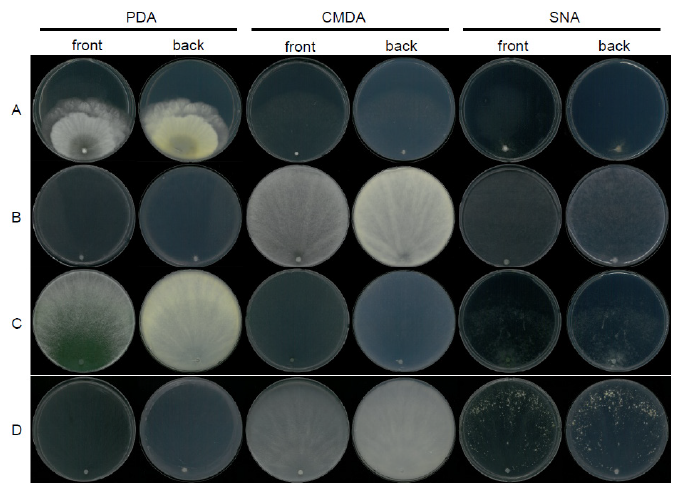
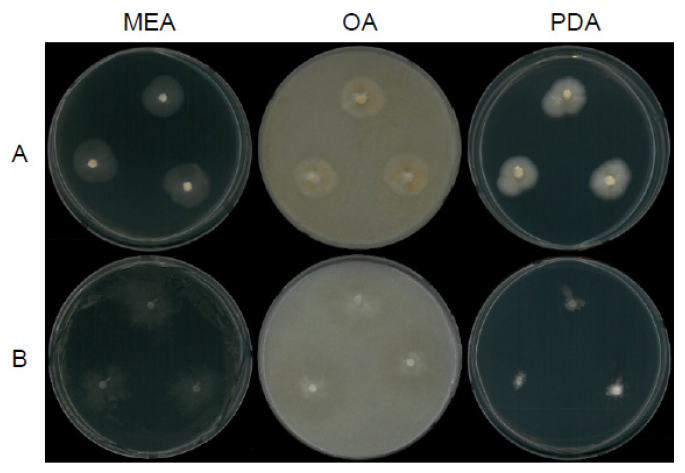
Fig. 12. Mycelial growth on potato dextrose agar (PDA), corn meal dextrose agar (CMDA), and synthetic nitrogen-poor or nutrient-poor agar (SNA) for 7 days at 25℃. A, Trichoderma appalachiense strain NNIBRFG15408; B, Trichoderma subviride strain NNIBRFG6329; C, Trichoderma taiwanense strain NNIBRFG20685; D, Trichoderma tsugarense strain NNIBRFG23468.

Fig. 13. Microscopic observation. A-B, conidiophore (A) and conidia (B) of Trichoderma appalachiense strain NNIBRFG15408; C-D, conidiophore (C) and conidia (D) of Trichoderma subviride strain NNIBRFG6329; E-F, conidiophore (E) and conidia (F) of Trichoderma taiwanense strain NNIBRFG20685; G-I, conidiophore (G), chlamydospore (H), and conidia (I) of Trichoderma tsugarense strain NNIBRFG23468. Scale bars indicate 10 μm (B, D, F, H-I) and 50 μm (A, C, E, G).
Trichoderma appalachiense Samuels & Jaklitsch, Persoonia 31: 135 (2013) [MB#803627] (Figs. 12A and 13A-B)
Description: Colonies grew slightly slowly at 25℃ and reached 56 mm on PDA, 64 mm on CMDA, and 16 mm on SNA, 7 d after inoculation. The colony on PDA was hyaline to white, forming concentric rings with a cottony aerial mycelial surface, and its reverse side was white to light yellow from the center. The colony on CMDA was hyaline with little aerial mycelia, and its reverse side was hyaline. The colony on SNA was hyaline with a little cottony aerial mycelial surface, and its reverse side was hyaline. The conidiophores branched divergently. Phialides were lageniform, arising directly from the main axis or terminating lateral branches, solitary or in clusters, and measured 7.1-11.8 µm×2.3-4.3 µm at the widest point (x=9.8±1.33 µm×3.2±0.45 μm, L/W ratio=3.03, n=30). Conidia were green, aseptate, ellipsoid or subglobose, and thick-walled, measuring 3.0-4.8 µm×2.7-4.0 μm (x=3.8±0.35 µm×3.3±0.27 μm, L/W ratio=1.14, n=50).
Habitat: Filtered freshwater from stream
Specimen examined: Sanghyo-dong, Seogwipo-si, Jeju-do, Republic of Korea, 19 March 2019, NNIBRFG15407, Nakdonggang National Institute of Biological Resources
Note: Trichoderma appalachiense was isolated from decorticated wood in the mid-Atlantic states of the USA [41]. In this study, NNIBRFG15407 was isolated from filtered freshwater from a stream in Jeju Island. Moreover, this strain exhibited weak siderophore production (Table 3).
Trichoderma subviride W.T. Qin & W.Y. Zhuang, Scientific Reports 6 (no. 27074): 11 (2016) [MB#570245] (Figs. 12B and 13C-D)
Description: Colonies grew fast at 25℃ and reached 87 mm on PDA, 87 mm on CMDA, and 87 mm on SNA, 4 d after inoculation. The colony on PDA was white to green, with a fluffy aerial mycelial surface, and its reverse side was white to light yellow. The colony on CMDA was hyaline with little aerial mycelia, and its reverse side was hyaline. The colony on SNA was hyaline with a little cottony aerial mycelial surface, and its reverse side was hyaline. The conidiophores branched divergently. Phialides were lageniform or ampulliform, arising directly from the main axis or terminating lateral branches, solitary or in clusters, measuring 4.3-12.5 µm×2.0-3.7 µm at the widest point (x=7.3±1.62 µm×2.8±0.42 μm, L/W ratio=2.56, n=30). Conidia were green, aseptate, subglobose or globose, and measured 3.0–4.5 µm × 2.8–3.9 μm (x=3.7±0.31 µm×3.4±0.27 μm, L/W ratio=1.07, n=50). Chlamydospores were aseptate, globose or subglobose, and thick-walled, measuring 7.0-11.2 µm×6.3-11.4 μm (x=9.3±1.31 µm×8.5±1.08 μm, L/W ratio=1.09, n=20).
Habitat: Sediment in freshwater
Specimen examined: Ihwa-ri, Ubo-myeon, Gunwi-gun, Gyeongsangbuk-do, Republic of Korea, 22 June 2018, NNIBRFG6329, Nakdonggsang National Institute of Biological Resources
Note: Trichoderma subviride was isolated from twigs in China [42]. In this study, NNIBRFG6329 was isolated from sediment in Wicheon. Moreover, this strain exhibited antifungal activity against Fusarium oxysporum and F. solani, extracellular laccase activity, and siderophore production (Table 3).
Trichoderma taiwanense Samuels & M.L. Wu, Studies in Mycology 56: 130 (2006) [MB#501048] (Figs. 12C and 13E-F)
Description: Colonies grew fast at 25℃ and reached 87 mm on PDA, 87 mm on CMDA, and 71 mm on SNA, 7 d after inoculation. The colony on PDA was white to dark green, with a fluffy aerial mycelial surface, and its reverse side was white to light yellow. The colony on CMDA was hyaline with little aerial mycelia, and its reverse side was hyaline. The colony on SNA was hyaline with a little cottony aerial mycelial surface, and its reverse side was hyaline. The conidiophores branched divergently. Phialides were lageniform, arising directly from the main axis or secondary branches, solitary or in clusters, and measured 5.1-13.0 µm×1.1-4.3 µm at the widest point (x=8.1±1.78 µm×2.0±0.70 μm, L/W ratio=4.01, n=20). Conidia were green, aseptate, and subglobose or globose, measuring 3.2-4.7 µm×2.4-4.1 μm (x=4.0±0.37 µm×3.1±0.28 μm, L/W ratio=1.29, n=50). Chlamydospores were not observed in this study.
Habitat: Sediment in freshwater
Specimen examined: Iyeon-ri, Danbuk-myeon, Uiseong-gun, Gyeongsangbuk-do, Republic of Korea, 05 December 2018, NNIBRFG20685, Nakdonggang National Institute of Biological Resources
Note: Trichoderma taiwanense was isolated from barks in Taiwan [43]. In this study, NNIBRFG20685 was isolated from sediment in Wicheon. This strain showed extracellular laccase activity and siderophore production (Table 3).
Trichoderma tsugarense Yabuki & Okuda, Mycoscience 55(3): 209 (2014) [#MB 804140] (Figs. 12D and 13G-I)
Description: Colonies grew fast at 25℃ and reached 87 mm on PDA, 87 mm on CMDA, and 87 mm on SNA, 7 d after inoculation. The colony on PDA was white to beige, with a fluffy aerial mycelial surface, and its reverse side was white. The colony on CMDA was hyaline with little aerial mycelia, and its reverse side was hyaline. The colony on SNA was hyaline with light yellow pustules in the margins and a little cottony aerial mycelial surface, and its reverse side was hyaline. Phialides were lageniform, solitary or in clusters, and measured 7.3-12.8 µm×2.3-3.9 µm at the widest point (x=9.4±1.32 µm×3.6±0.59 μm, L/W ratio=2.63, n=20). Conidia were green, aseptate, and subglobose or globose, measuring 4.1-6.6 µm×2.5-5.2 μm (x=5.3±0.58 µm×3.8±0.68 μm, L/W ratio=1.40, n=50). Chlamydospores were aseptate, globose or subglobose, and thick-walled, measuring 7.7-11.6 µm×6.6-9.2 μm (x=9.0±1.51 µm×7.6±0.99 μm, L/W ratio=1.18, n=20).
Habitat: Sediment in freshwater
Specimen examined: Iyeon-ri, Danbuk-myeon, Uiseong-gun, Gyeongsangbuk-do, Republic of Korea, 05 December 2018, NNIBRFG20685, Nakdonggang National Institute of Biological Resources
Note: Trichoderma tsugarense was isolated from volcanic ash soil in Japan [44]. In this study, NNIBRFG23468 was isolated from sediment in Wicheon. This strain exhibited extracellular laccase activity and siderophore production (Table 3).