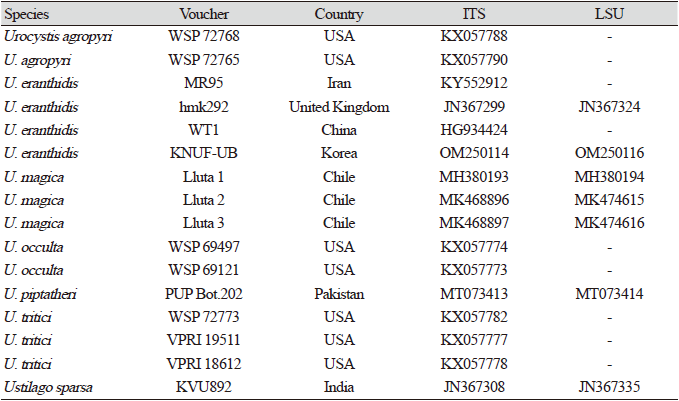
The genus Anemone (family Ranunculaceae) has more than 150 species of flowering plants distributed globally, but is native to temperate zones of both northern and southern hemispheres [1,2]. Anemone flaccida (Flaccid Anemone), a medicinal perennial grass distributed globally, has been used to traditionally treat rheumatism and neuralgia and has reported anticonvulsant, anti-inflammatory, anticancer, immunosuppressive, and antiviral activities [1,3,4]. In Korea, A. flaccida is located in southern regions such as Jeolla, Gyeongnam provinces, and Jeju Island, and has a high academic and horticultural value [5,6]. Previous studies have reported A. flaccida as a plant host for pathogens such as Puccinia japonica, Ceraceopsora elaeagni, and Urocystis pseudoAnemones [7-9], the last one reported in Japan. However, no study exists about the disease occurrence on A. flaccida in Korea so far.
In April 2021, abnormal spots were observed on leaves and stems of A. flaccida at the historical landmark of Ban-Gu Jeong, the native habitat of A. flaccida in Haman, Gyeongnam province, Korea. The symptomatic areas presented gray-colored swellings or blisters on leaves and stems, containing a black powdery mass of spores (Figs. 1A-E). The morphological characteristics of the designated KNUF-UB strain were observed through light microscopy using a BX50 microscope (Olympus, Tokyo, Japan), and the spores were measured using Olympus cellSens imaging software. Sori in leaves presented irregular, globose spore balls of 21.96-39.36×15.75-22.71 µm consisting of 1-3 spores, surrounded by a layer of pale yellow, sometimes collapsed sterile cells (Figs. 1F-I). The observed symptoms matched previous Urocystis eranthidis reports [10,11], and the aforementioned morphological characteristics of KNUF-UB isolate agreed well with those previously recorded for U. eranthidis [12,13].
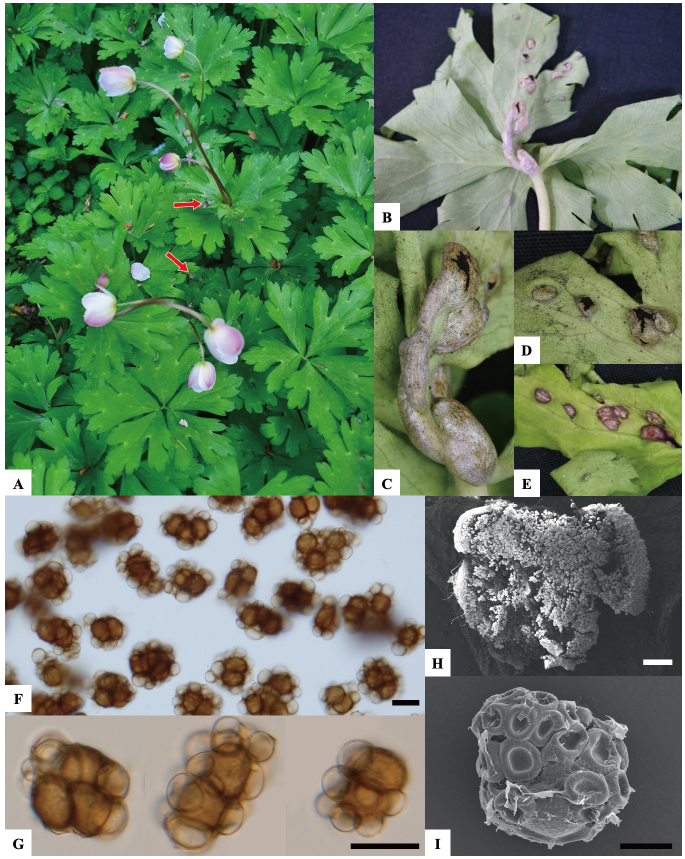
Fig. 1. Symptoms observed on Anemone flaccida and morphological characteristics of the KNUFUB isolate. (A) Abnormal spots on leaves and stems. (B-E) Blisters containing a black powdery mass of spores. (F and G) Light microscopy of spore balls (H) SEM of spore balls (I) SEM of spore ball with collapsed sterile cells around. Red arrow: Natural symptoms on the host leaf; Black scale bar: 10 μm; White scale bar: 200 μm.
To identify the KNUF-UB strain, genomic DNA was extracted using a HiGene Genomic DNA Prep Kit (BIOFACT, Daejeon, Korea) following the manufacturer's protocol. Identification was conducted using the nuclear ribosomal internal transcribed spacer (ITS) region and the large subunit (LSU) ribosomal RNA gene [14]. For the ITS region, ITS1F/ITS4 [15,16] primer pair was used, and the LR0R/LR6 [17] primer pair was used for the partial LSU gene. Amplified PCR products were purified using ExoSAP-IT PCR Product Cleaning Reagent (Thermo Fisher Scientific, Waltham, MA, USA) and sequenced (Bioneer Co., Daejon, Korea). Sequences of 664 and 1,031bp were obtained from the ITS region and partial LSU gene, respectively. All obtained gene sequences were registered in the National Center for Biotechnology Information (NCBI) with the following GenBank accession numbers (OM250114, OM250116). A BLAST search in NCBI’s database revealed that KNUF-UB had a 98.2% and 97.9% similarity with U. eranthidis MR95 (KY552912) and U. eranthidis hmk292 (JN367299) for the ITS region, respectively. For the LSU gene, KNUF-UB showed 99.8% similarity with U. eranthidis hmk292 (JN367324) and U. eranthidis M0065996 (EF517940). To identify the phylogenetic relationships, Urocystis spp. sequences were retrieved from NCBI's GenBank (Table 1), and a neighbor-joining phylogenetic analysis for each genetic marker was conducted using MEGA version X [18]. Consequently, the KNUF-UB strain was clustered together with U. eranthidis hmk292 in a clade with a high bootstrap value for both the ITS region and LSU gene (Figs. 2 and 3), strongly supporting the affiliation to the same species.
Urocystis eranthidis is a typical spring smut fungus first described in 1950 [19]. Smut fungi are pathogens that occur worldwide and mainly infect grasses by producing sori in various host plant organs [20,21]. The genus Urocystis (order Urocystidales, family Urocystidaceae) contains more than 170 species of smut fungi pathogenic to plants [22]. For the host plant family of Ranunculaceae, Urocystis can be observed causing sori on leaves, with spore balls of one to many fertile spores [11]. In Anemone species, U. anemones, U. japonica, U. antipolitana, and U. pseudoanemones, have been previously reported [9]. U. eranthidis has been reported as a pathogen for Ceratocephalus falcatus and Eranthis hymenalis [23-25]. However, it has not been previously reported on Anemone spp. In Korea, only three species of the genus Urocystis have been reported; U. colchici, U. syncocca, and U. tritici [26]. To the best of our knowledge, this is the first report of smut caused by U. eranthidis on A. flaccida in Korea, highlighting the importance of disease incidence research to understand and control this and other potential pathogens.er

Fig. 2. Molecular phylogenetic analysis of internal transcribed spacer (ITS) region sequences of genus Urocystis inferred using the neighbor-joining method. The percentage in which the associated taxa clustered together is shown next to the branches. The tree is drawn to scale, with branch lengths measured in the number of substitutions per site. Strain is shown in bold, and values lower than 70 are not shown.

Fig. 3. Molecular phylogenetic analysis of large subunit (LSU) rRNA gene sequences of genus Urocystis inferred using the neighbor-joining method. The percentage in which the associated taxa clustered together is shown next to the branches. The tree is drawn to scale, with branch lengths measured in the number of substitutions per site. Strain is shown in bold, and values lower than 70 are not shown.
Acknowledgments
This work was supported by the Korea Institute of Planning and Evaluation for Technology in Food, Agriculture and Forestry (IPET) through the Agriculture, Food and Rural Affairs Convergence Technologies Program for Educating Creative Global Leader Program funded by the Ministry of Agriculture, Food and Rural Affairs (MAFRA) (no. 321001-03).