Korean Journal of Mycology (Kor J Mycol) 2023 December, Volume 51, Issue 4, pages 490. https://doi.org/10.4489/KJM.20230048
Received on November 06, 2023, Revised on December 26, 2023, Accepted on December 26, 2023.
Copyright © The Korean Society of Mycology.
This is an Open Access article which is freely available under the Creative Commons Attribution-NonCommercial 4.0 International License (CC BY-NC) (https://creativecommons.org/licenses/by-nc/4.0/).
INTRODUCTION
Since long time, apples and pears have been in the spotlight as internationally important crops to the extent that more than 100 million tons of apples and pears are produced worldwide annually [1-3]. In 2020, Korea produced approximately 550, 000 tons of apples and pears. Soil is indispensable for growing apple and pear trees. Soil supports the roots of trees and stores moisture; various interactions, such as helping or hindering the growth of trees by various rhizosphere microorganisms, occur in the soil [4,5].
Diverse fungi infect apple and pear trees [6]. Orchard soil serves as a reservoir for these pathogenic fungi, as they can overwinter in soil. We investigated fungi from apple and pear trees and orchard soils in diverse places to monitor diseases. Diverse fungi, including pathogenic fungi, were identified. Among the identified fungi, seven species belonging to the phylum Ascomycota have not been recorded in Korea. In this paper, we report the full descriptions and provide illustrations of their morphological characteristics and molecular phylogenetic positions.
MATERIALS AND METHODS
Collecting rhizosphere soil samples: The rhizosphere soils of apple and pear trees were collected from orchards in Cheongju, Anseong, and Cheonan, Korea. After excavating approximately 30 cm from the surface, the soil (approximately 100 g) around the roots of apple and pear trees was collected using a spade and collected in a 50 mL conical tube. Soil-containing tubes were stored in a cooler and transported to the laboratory for fungal isolation.
Isolation of fungi: Further, 3 g of the collected soil was placed in a 50 mL conical tube containing 10 mL of sterile distilled water and vortexed for 30 min. The mixture was then diluted up to 1,000 times through serial dilution; 1 mL of muddy water was mixed with 9 mL of sterile distilled water. Diluted muddy water (100 µL) was dispensed onto Dichloran-Glycerol 18% (DG18) agar medium and spread using a glass spreader. DG18 agar plates spread with soil dilution were incubated for at least three weeks in an incubator at 25℃. Among the growing fungal mycelia, mycelia that appeared different in morphology were collected and separated in pure form on potato dextrose agar (PDA). Purely isolated strains were cultured on PDA, malt extract agar (MEA), czapek yeast extract agar (CYA), and oatmeal agar (OA) to observe colony morphology.
Microstructure observation: The microstructures of the isolated fungi were observed using an optical microscope (BX53; Olympus, Tokyo, Japan). The size of each microstructure was measured 50 times, with the range shown.
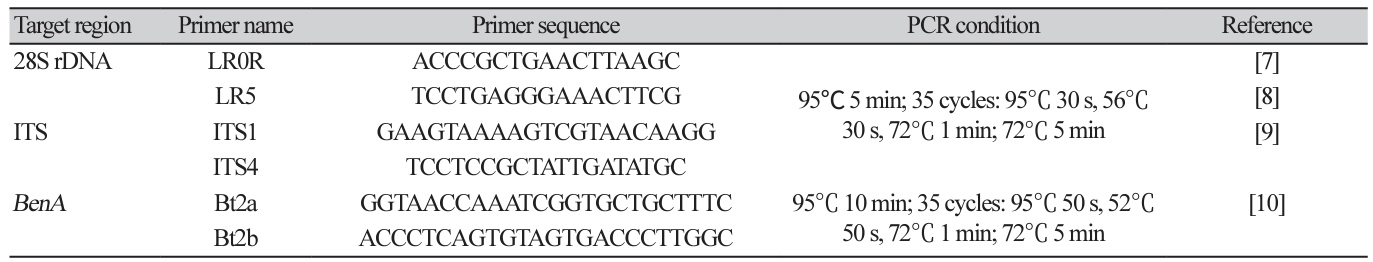
DNA extraction and phylogenetic analysis: Purely isolated fungi were cultured on PDA lined with sterilized cellophane for another week and approximately 50 mg of mycelia was scraped with a surgical blade and collected in a 2 mL tube. DNA was extracted from the collected mycelia using a Plant/Fungi DNA Isolation Kit (Norgen Biotek Corp., Thorold, Canada). Using the extracted DNA as a template, PCR primers in Table 1 were used to amplify the internal transcribed spacer (ITS) region (ITS1-5.8S-ITS2), the large subunit of nuclear ribosomal RNA (28S rDNA), and partial β-tubulin gene (BenA) sequences. PCR was performed under the conditions listed in Table 1 using a Bio-Rad T100 Thermal cycler. After confirming PCR product amplification by electrophoresis on a 0.8% agarose gel, the PCR amplicon was purified and sequenced by Macrogen Corp. (Seoul, Korea).
The determined nucleotide sequences were analyzed for homology using the National Center for Biotechnology Information (NCBI, https://www.ncbi.nlm.nih.gov/) search engine GenBank BLASTN. Fungal species closely related to the taxa isolated in this study and belonging to the same or different genera, were included in the analysis. Reference sequences were obtained from GenBank and are listed in Tables 2-8. Individual sequence datasets of the ITS region, 28S rDNA, and the β-tubulin gene were aligned using the MUSCLE (multiple Sequence Comparison by Log- Expectation) alignment tool of MEGA XI [11] and improved manually where necessary using BioEdit [12]. Based on the aligned sequences, a maximum likelihood (ML) phylogenetic tree was constructed and the reliability of each node in the phylogenetic tree was evaluated using 1,000 bootstraps [13].
Table 1 Primer information and PCR amplification condition used to amplify each target region
RESULTS AND DISCUSSION
The morphological characteristics of each species of unrecorded soil fungi isolated and identified in this study and the results of phylogenetic analysis based on nucleotide sequence analysis of ITS and 28S rDNA are presented below. The seven DUCC strains were deposited at the National Institute of Biological Resources of the Republic of Korea (https://www.nibr.go.kr/cmn/main/enMain.do) and received the NIBRFGC numbers. The analyzed nucleotide sequences were registered in GenBank of the NCBI database and the registration numbers of the seven DUCC strains are provided in Tables 2-8.
REFERENCE
D O'Rourke A. The world apple market. London: Routledge; 2018.
https://doi.org/10.1201/9780203719091
Deckers T, Schoofs H. Status of the pear production in Europe. In X Int Pear Symposium 2007;800:95-106.
https://doi.org/10.17660/ActaHortic.2008.800.8
Teng Y. The pear industry and research in China. Acta Hortic 2011;909:161-70.
https://doi.org/10.17660/ActaHortic.2011.909.16
Mazzola M, Hewavitharana SS, Strauss SL. Brassica seed meal soil amendments transform the rhizosphere microbiome and improve apple production through resistance to pathogen reinfestation. Phytopathology 2015;105:460-9.
https://doi.org/10.1094/PHYTO-09-14-0247-R
Van Horn C, Somera TS, Mazzola M. Comparative analysis of the rhizosphere and endophytic microbiomes across apple rootstock genotypes in replant orchard soils. Phytobiomes J 2021;5:231-43.
https://doi.org/10.1094/PBIOMES-08-20-0058-R
Sutton TB, Aldwinckle HS, Agnello AM, Walgenbach JF. Compendium of apple and pear diseases and pests. Second Edition. St. Paul: APS Publications; 2014.
https://doi.org/10.1094/9780890544334
Cubeta MA, Echandi E, Abernethy T, Vilgalys R. Characterization of anastomosis groups of binucleate Rhizoctonia species using restriction analysis of an amplified ribosomal RNA gene. Phytopathology 1991;81:1395-400.
https://doi.org/10.1094/Phyto-81-1395
Vilgalys R, Hester M. Rapid genetic identification and mapping of enzymatically amplified ribosomal DNA from several Cryptococcus species. J Bacteriol 1990;172:4238-46.
https://doi.org/10.1128/jb.172.8.4238-4246.1990
White TJ, Bruns T, Lee SJWT, Taylor J. Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics. PCR Methods Appl 1990;18:315-22.
https://doi.org/10.1016/B978-0-12-372180-8.50042-1
Glass NL, Donaldson GC. Development of primer sets designed for use with the PCR to amplify conserved genes from filamentous ascomycetes. Appl Environ Microbiol 1995;61:1323-30.
https://doi.org/10.1128/aem.61.4.1323-1330.1995
Tamura K, Stecher G, Kumar S. MEGA11: molecular evolutionary genetics analysis version 11. Mol Biol Evol 2021;38:3022-7.
https://doi.org/10.1093/molbev/msab120
Hall TA. BioEdit: a user friendly biological sequence alignment editor and analysis program for Window 95/98/NT. Nucleic Acids Symp Ser 1999;44:95-8.
Thorne JL, Kishino H, Felsenstein J. An evolutionary model for maximum likelihood alignment of DNA sequences. J Mol Evol 1991;33:114-24.
https://doi.org/10.1007/BF02193625
Trakunyingcharoen T, Lombard L, Groenewald JZ, Cheewangkoon R, To-anun C, Alfenas AC, Crous PW. Mycoparasitic species of Sphaerellopsis and allied lichenicolous and other genera. IMA Fungus 2014;5:391-414.
https://doi.org/10.5598/imafungus.2014.05.02.05
Shoemaker RA, Babcock CE, Irwin JAG. Massarina walkeri nov. sp., the teleomorph of Acrocalymma medicaginis from Medicago sativa contrasted with Leptosphaeria pratensis, L. weimeri n. sp., and L. viridella. Can J Bot 1991;69:569-73.
https://doi.org/10.1139/b91-077
Lai D, Wang A, Cao Y, Zhou K, Mao Z, Dong X, Tian J, Xu D, Dai J, Peng Y, et al. Bioactive dibenzo-α-pyrone derivatives from the endophytic fungus Rhizopycnis vagum Nitaf22. J Nat Prod 2016;79:2022-31.
https://doi.org/10.1021/acs.jnatprod.6b00327
He C, Wang W, Hou J. Plant growth and soil microbial impacts of enhancing licorice with inoculating dark septate endophytes under drought stress. Front Microbiol 2019;10:2277.
https://doi.org/10.3389/fmicb.2019.02277
Tibpromma S, Hyde KD, McKenzie EHC, Bhat DJ, Phillips AJL, Wanasinghe DN, Samarakoon MC, Jayawardena RS, Dissanayake AJ, Tennakoon DS, et al. Fungal diversity notes 840-928: Micro-fungi associated with Pandanaceae. Fungal Divers 2018;93:1-160.
https://doi.org/10.1007/s13225-018-0408-6
Abang MM, Kabbabeh S, Ahmed S, Murad S, Chilvers MI, Peever TL, Schroers HJ. First report of chickpea wilt caused by Clonostachys rhizophaga in Syria. Plant Dis 2009;93:666.
https://doi.org/10.1094/PDIS-93-6-0666A
Sun ZB, Li SD, Ren Q, Xu JL, Lu X, Sun MH. Biology and applications of Clonostachys rosea. J Appl Microbiol 2020;129:486-95.
https://doi.org/10.1111/jam.14625
Chethana KWT, Zhou Y, Zhang W, Liu M, Xing QK, Li XH, Yan JY, Chethana KWT, Hyde KD. Coniella vitis sp. nov. is the common pathogen of white rot in Chinese vineyards. Plant Dis 2017;101:2123-36.
https://doi.org/10.1094/PDIS-12-16-1741-RE
Yin X, Li T, Jiang X, Tang X, Zhang J, Yuan L, Wei Y. Suppression of grape white rot caused by Coniella vitis using the potential biocontrol agent Bacillus velezensis GSBZ09. Pathogens 2022;11:248.
https://doi.org/10.3390/pathogens11020248
Rossman AY, Samuels GJ, Rogerson CT, Lowen R. Genera of Bionectriaceae, Hypocreaceae and Nectriaceae (Hypocreales, Ascomycetes) (No. 42). Stud Mycol 1999;42:1-248.
Herrera CS, Hirooka Y, Chaverri P. Pseudocospeciation of the mycoparasite Cosmospora with their fungal hosts. Ecol Evol 2016;6:1504-14.
https://doi.org/10.1002/ece3.1967
Malloch D, Cain RF. Four new genera of cleistothecial Ascomycetes with hyaline ascospores. Can J Bot 1971;49:847-54.
https://doi.org/10.1139/b71-125
Crous PW, Cowan DA, Maggs-Kölling G, Yilmaz N, Larsson E, Angelini C, Brandrud TE, Dearnaley JDW, Dima B, Dovana F, et al. Fungal planet description sheets: 1112-1181. Persoonia 2020;45:251-409.
https://doi.org/10.3767/persoonia.2020.45.10
Babu AG, Kim SW, Yadav DR, Hyum U, Adhikari M, Lee YS. Penicillium menonorum: a novel fungus to promote growth and nutrient management in cucumber plants. Mycobiology 2015;43:49-56.
https://doi.org/10.5941/MYCO.2015.43.1.49
Wang XW, Bai FY, Bensch K, Meijer M, Sun BD, Han YF, Crous PW, Samson RA, Yang FY, Houbraken J. Phylogenetic re-evaluation of Thielavia with the introduction of a new family Podosporaceae. Stud Mycol 2019;93:155-252.
https://doi.org/10.1016/j.simyco.2019.08.002
Sishuba A. Biodiversity and antimicrobial activity of endophytic fungi isolated from native Sutherlandia frutescns (cancer bush) [dissertation]. Potchefstroom: North West University; 2022.
Fernandes MLP, Bastida F, Jehmlich N, Martinović T, Větrovský T, Baldrian P, DelgadoBaquerizo M, Starke R. Functional soil mycobiome across ecosystems. J Proteome 2022;252:104428.
https://doi.org/10.1016/j.jprot.2021.104428
Vétrovský T, Kohout P, Kopecký M, Machac A, Man M, Bahnmann BD, Brabcová V, Choi J, Meszárošová L, Human ZR, et al. A meta-analysis of global fungal distribution reveals climate-driven patterns. Nat Commun 2029:10:5142.
https://doi.org/10.1038/s41467-019-13164-8
Põlme S, Abarenkov K, Nilsson HR, Lindahl BD, Clemmensen KE, Kauserud H, Nguyen N, Kjøller R, Bates ST, Baldrian P, et al. Fungal traits: a user-friendly traits database of fungi and fungus-like stramenopiles. Fungal Divers 2020:105:1-16.

