Jun-Woo Choi1, Seong-Keun Lim1, Seo-Ryeong Lee1, Chang-Gi Back2, In-Kyu Kang3, Seung-Yeol Lee1,4,*, Hee-Young Jung1,4
1Department of Plant Medicine, Kyungpook National University, Daegu 41566, Korea
2Department of Environmental Horticulture and Landscape Architecture, Environmental Horticulture, Dankook University, Cheonan 31116, Korea
3Department of Horticultural Science, Kyungpook National University, Daegu 41566, Korea
4Institute of Plant Medicine, Kyungpook National University, Daegu 41566, Korea
*Correspondence to leesy1123@knu.ac.kr
Korean Journal of Mycology (Kor J Mycol) 2024 June, Volume 52, Issue 2, pages 145-153. https://doi.org/10.4489/kjm.520207
Received on June 04, 2024, Revised on June 22, 2024, Accepted on June 24, 2024, Published on June 30, 2024.
Copyright © The Korean Society of Mycology.
This is an Open Access article which is freely available under the Creative Commons Attribution-NonCommercial 4.0 International License (CC BY-NC) (https://creativecommons.org/licenses/by-nc/4.0/).
While investigation of the fungal diseases on apples collected from Cheongsong-gun and Bonghwa-gun in Gyeongbuk province, Korea, between August and September 2023 isolated f ive fungal strains from fruits with sooty blotch and flyspeck (SBFS) disease. The strains were designated as KNUF-23-CS02, KNUF-23-CS-06, KNUF-23-CS12, KNUF-23-BH01, and KNUF-23BH03. When grown on potato dextrose agar and 2% water agar, the cultural characteristics of the strains were similar to those previously reported characteristics of Peltaster fructicola Pf001. The strains produced monoblastic, hyaline conidiogenous cells; the conidia were hyaline, unicellular, cylindrical to ovoidal, and 3.5-7×1.7-3.9 and 4.0-6.6×1.8-3.2 μm in size on synthetic nutrient-poor agar or water agars, respectively. Secondary conidia production by microcyclic conidiation and budding was observed. The KNUF-23-BH03 strain was shown to cause SBFS symptoms similar to those observed on the apples in the pathogenicity test. Molecular phylogenetic analyses were conducted based on the isolated species sequences of the internal transcribed spacer region, nuclear large ribosomal DNA subunit, and mitochondrial small ribosomal RNA subunit gene. The five strains were clustered with Peltaster fructicola Pf001. Based on the cultural and morphological characteristics and phylogenetic analysis, the five strains were identified as Peltaster fructicola, which has not been previously reported in Korea.
Apple, Morphology, Peltaster fructicola, Phylogenetic analysis, Sooty blotch and flyspeck disease
Apples (Malus domestica) are an important crop in Korea, which produced 394,428 tons of fruit and cultivated 33,789 ha in 2023, of which 59% of the cultivated regions were located in the Gyeongbuk province [1]. Fungal diseases are a primary cause of reduced fruit quality and yield in apple production [2]. For example, fungal species that cause sooty blotch and flyspeck (SBFS) colonize the epicuticular wax layer of apple fruits; this causes no physiological damage to the living cells [3]. The blemishes on apple fruits surfaces are caused by the mycelial mat and sclerotium-like body of the SBFS-causing species and can decrease their fresh market value by up to 90% [4]. In addition, these fungal species can accelerate the apples weight loss and shriveling during cold storage [5]. Therefore, SBFS-causing fungi are economically significant pathogens in apples worldwide. Globally, more than 100 species have been recorded to cause SBFS, including species from the genera Peltaster, Schizothyrium, Ramularia, and Cyphellophora [6]. The genus Peltaster was established by Sydow and Sydow (1917) based on P. hedyotidis Syd. & P. Syd. isolated from Hedyotis elmeri (Rubiaceae) in the Philippines [7]. In 1996, P. fructicola was first described as causing SBFS with a punctate mycelial type on apple fruits and the stems of brambles (Rubus spp.) in the United States of America (USA) [8,9]. Subsequently, P. cerophilus and P. gemmifer have been associated with causing SBFS on apples in Slovenia and USA, respectively [7,10]. In Korea, two species, Gloeodes pomigena and Schizothyrium pomi, have been reported as SBFS causal agents on apples [11]; however, there are no previously reported studies on the genus Peltaster species that may be associated with SBFS on apples in Korea. Therefore, apple fruits showing symptoms of SBFS were collected from apple orchards in Cheongsong-gun and Bonghwa-gun in the Gyeongbuk province of Korea in 2023. In this study, we report unrecorded pathogen that caused SBFS on apples in Korea based on its morphological characteristics and phylogenetic analysis.
Apple fruits (cv. Fuji) with SBFS disease symptoms were collected from apple orchards located in Cheongsong-gun (36°17′09.6″N 128°57′30.9″E) and Bonghwa-gun (36°54′09.4″N 128°58′14.5″E) in the Gyeongbuk province of South Korea between August and September 2023. The disease appeared as dark regular or irregular sclerotium-like bodies with mycelial mats on the apple surfaces. The causal agents of the SBFS were isolated from symptomatic apples according to the method of Medjedović et al. [10]. First, the apple surfaces were sterilized with 70% ethanol. Then, the presumptive causative agents of the SBFS were transferred from the apple peels to potato dextrose agar (PDA) (Difco™, Becton, Dickinson and Company [BD], Franklin Lakes, NJ, USA) and cultured at 25℃ in the dark. After 2 weeks, small black colonies were observed, and the margin of each colony was transferred to new PDA plates. The five isolated strains were designated as KNUF-23-BH01, KNUF-23-BH03, KNUF-23-CS02, KNUF-23-CS06, and KNUF-23CS12; strain KNUF-23-BH03 was selected for further morphological and cultural characterization.
Cultural characteristics of the KNUF-23-BH03 were observed after growth on PDA and 2% water agar (WA) following the method of Williamson et al. [9]. Mycelial plugs were taken from the margin of the colony using a 4 mm cork borer, placed on the PDA or 2% WA, and then incubated at 25℃ in the dark for 3 weeks. Morphological characteristics were observed using synthetic nutrient-poor agar (SNA) and 2% WA according to a previously reported method [12]. The conidia and conidiogenous cells of the fungal specimens were observed and measured using an optical microscope (Olympus BX-50; Evident Corp., Tokyo, Japan). The diameter of the colonies was measured with vernier calipers (Mitutoyo Corp., Kawasaki, Japan).
The total genomic DNA was extracted from the fungal strain KNUF-23-BH03 grown on the PDA using a HiGene™ Genomic DNA Prep Kit (Biofact Co., Ltd., Daejeon, Korea) according to the manufacturer’s protocol. For molecular identification of the strain, the internal transcribed spacer (ITS) region, nuclear large subunit (LSU) ribosomal DNA region, and mitochondrial small subunit (mtSSU) ribosomal DNA region were amplified with primer pairs ITS1F/ITS4, LROR/LR5, and mrSSU1/mrSSU3R [13- 17], respectively. The amplified PCR products were purified using ExoSAP-IT (Thermo Fisher Scientific Inc., Waltham, MA, USA) and sequenced by Solgent Co., Ltd. (Daejeon, Korea). All the obtained sequences were registered in GenBank as PP814955–PP814959 for the ITS region, PP814967-PP814971 for LSU, and PP824772PP824776 for mtSSU.
The phylogenetic analysis was conducted using retrieved sequences registered on the NCBI (Table 1). The ambiguous regions were deleted from the alignments and the evolutionary distance matrices for the maximum likelihood (ML) method were calculated using the Clustal X program and Tamura-Nei model [18]. The ML method was used for the construction of phylogenetic trees MEGA 11.0 software with bootstrap values based on 1,000 replications [19].
Table 1. GenBank accession numbers of the Peltaster species used for phylogenetic analyses
| Species | Strain number | Accession number | |||
|---|---|---|---|---|---|
| Country | ITS | LSU | mtSSU | ||
| Capnodium coffeae | CBS 147.52 | N/A | DQ491515 | DQ247800 | FJ190609 |
| Peltaster cerophilus | 11151 | Slovenia | JN573672 | JN573664 | KF550949 |
| Peltaster cerophilus | 318-07 | Breže, Slovenia | JN573676 | JN573662 | KF550952 |
| Peltaster fructicola | 11157 | Slovenia | JN573670 | JN573665 | KF550948 |
| Peltaster fructicola | LNHT1506 | China | JX961608 | KT780342 | KT780343 |
| Peltaster fructicola | Pf001 | North Carolina, USA | MF075296 | AY598927 | MF075289 |
| Peltaster fructicola | KNUF-23-BH01 | Bonghwa, Korea | PP814956 | PP814970 | PP824772 |
| Peltaster fructicola | KNUF-23-BH03 | Bonghwa, Korea | PP814955 | PP814971 | PP824776 |
| Peltaster fructicola | KNUF-23-CS02 | Cheongsong, Korea | PP814959 | PP814969 | PP824775 |
| Peltaster fructicola | KNUF-23-CS06 | Cheongsong, Korea | PP814958 | PP814968 | PP824774 |
| Peltaster fructicola | KNUF-23-CS12 | Cheongsong, Korea | PP814957 | PP814967 | PP824773 |
| Peltaster gemmifer | GTE9a | Illinois, USA | KF646814 | AY598929 | KF550946 |
| Peltaster gemmifer | UIE11b | Illinois, USA | AY598890 | KF550919 | KF550945 |
| Peltaster sp. | 65rap | Germany | JN573668 | DQ363413 | KF550947 |
| Peltaster sp. | KY3_8E1a | Kentucky, USA | FJ438384 | FJ147171 | KF550944 |
The strains identified in this study are indicated in bold.
N/A: Not available; ITS: Internal transcribed spacer regions; LSU: Nuclear large subunit ribosomal DNA region; mtSSU: Mitochondrial small subunit ribosomal DNA region.
Pathogenicity test was conducted to verify Koch’s postulates by inoculation of the KNUF-23-BH03 strain on healthy apple fruits (cv. Fuji) using a modification of the method mentioned by Johnson et al. [20]. First, the apple fruits were washed using tap water, surface sterilized with 70% ethanol, and then dried for 2 min. An inoculum suspension was prepared by homogenizing 0.05 g of mycelial fragments grown on PDA for 2 weeks with 1 mL of double-distilled water; the suspension was supplemented with 0.1% Tween 20. Then, sterilized paper discs that had been dipped in the inoculum suspension for 2 min were attached to the surface of the healthy apples. Inoculated fruits were incubated in a moist chamber at 25℃ for 3 weeks.
The mycological characteristics and molecular phylogeny of the five strains KNUF-23-BH01, KNUF23-BH03, KNUF-23-CS02, KNUF-23-CS06, and KNUF-23-CS12 isolated in this study were similar to each other. Therefore, the cultural and morphological characteristics of the KNUF-23-BH03 were described as the representative strain in this study.
The KNUF-23-BH03 showed a punctate mycelial type on the apple peel, including a visible mycelial mat with darkly pigmented, circular, or irregular sclerotium-like bodies, 65-163 μm diameter (n=50) (Fig. 1A-C). Previously, the sclerotium-like bodies of P. fructicola LNHT1506 and Pf001 were reported to be 68-145 μm and 19-282 μm in diameter, respectively [9,12]. The colony of KNUF-23-BH03 reached 1217 mm (av.=14 mm) in diameter after incubation at 25℃ for 3 weeks on PDA. The colonies, which were slow-growing, erumpent, dense, irregular with smooth margins, wrinkled surfaces, and had no aerial mycelium; in addition, primarily yellowish conidia masses that were reverse turned dark beige to olive-gray (Fig. 1). The colony of P. fructicola LNHT1506 and Pf001 grown on PDA have been previously reported as 13-16 mm (av.=14 mm) and 11-23 mm (av.=14 mm) in diameter, respectively; P. fructicola Pf001’s colonies have been reported as 8-22 mm (av.=14 mm) in diameter when cultivated on WA [9,12].
Furthermore, conidiophores were absent in the KNUF-23-BH03 and the hyphae were septate, branched, hyaline, and turned brown in older cultures. The conidiogenous cells were monoblastic, hyaline, and straightly formed from the hypha. The conidia were abundant, hyaline, solitary, unicellular, cylindrical to ovoidal, and 3.5-7.0×1.7-3.9 μm in size on SNA (n=50). In contrast, the conidia were slightly smaller (4.06.6×1.8-3.2 μm) on WA compared with on SNA. The conidia of P. fructicola LNH1506 were previously reported to be 4.0-6.5×1.5-2.5 μm and 4.0-6.0×1.5-2.0 μm in size on SNA and WA, respectively. The conidia size of P. fructicola Pf001 was 3.2-7.1×1.1-2.4 μm on WA [9,12]. In addition, the formations of secondary conidia by microcyclic conidiation and budding were observed (Fig. 1H-J). Microcyclic conidiation induced by environmental agents has been previously observed in other SBFS-causing fungi [21,22]. In addition, the production of secondary conidia through budding can be source of spreading, that previously reported on other Peltaster species [7]. The morphological and cultural characteristics of the strain KNUF-23-BH03 were similar to those of previously identified P. fructicola (Table 2) [9,12].
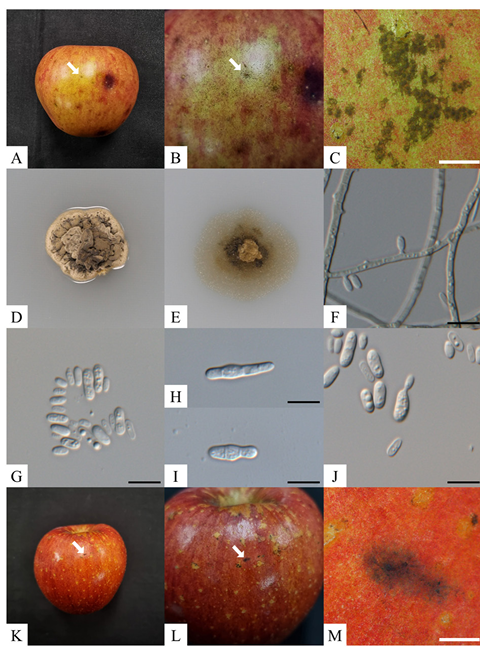
Fig. 1. Peltaster fructicola KNUF-23-BH03. A-C: Punctate symptoms on apple fruit; D, E: Enlarged colony grown on potato dextrose agar and 2% water agar, respectively, for 21 days at 25℃ in the dark; F: Conidiogenous cells; G: Conidia; H, I: Microcyclic conidiation; J: Primary conidia producing secondary conidia by budding. K-M: Results of the pathogenicity test. White scale bars=1 mm, black scale bars=10 μm.
Table 2. Morphological characteristics of the strain KNUF-23-BH03 compared with those previously reported for Peltaster species
| Characteristic | Peltaster fructicola KNUF-23-BH03a | Peltaster fructicola Pf001b | Peltaster fructicola LNHT1506c | Peltaster gemmifer GTE9ad |
|---|---|---|---|---|
| Colony | PDA: erumpent, wrinkled surface, with smooth margin, dark beige to olive-gray, reverse dark with yellow masses of conidia, 12-17 mm diam. after 21 days at 25℃ in the dark WA: flatten, circular, smooth margin, pale brown to dark brown, 14-20 mm diam. after 21 days at 25℃ in the dark | PDA: dense, compact, wrinkled or buckled, light gray, dark gray, dark brown or black, reverse dark brown with yellowgreen masses of conidia, 11-23 mm diam. after 21 days at 25℃ in the dark WA: indeterminate margin, white to pale brown, 8-22 mm diam. after 21 days at 25℃ in the dark | PDA: erumpent, with smooth, crenate margin, greenish gray to oliv aceousgray, reverse black with yellow masses of conidia at the bottom of the colony, 13-16 mm diam. after 21 days at 25℃ in the dark WA: undescribed | PDA: smooth edge, wrinkled surface, slightly raised, dark greenish olive, 15-16 mm diam. after 1 month at 25℃ WA: undescribed |
| Conidiophore | Absence | Absence | Absence | Single or branching conidiophores bearing primary conidia 19-45×1-1.5 μm |
| Conidia | Hyaline, solitary, unicellular, cylindrical to ovoidal, 3.5-7×1.7-3.9 μm on SNA and 4.0- 6.6×1.8-3.2 μm on WA | Unicellular, hyaline, ellipsoidal to ovoidal, 3.2-7.1×1.1-2.4 μm on WA | Solitary, unicellular, hyaline, elliptic to ovoidal, 4.0-6.5×1.5- 2.5 μm on SNA and 4.0 -6.0×1.5-2.0 μm on WA | Unicellular, cylindrical, 3.0 -6.0×1.5-4.0 μm on SNA |
| Mycelial type | Punctate, visible mycelial mat with darkly pigmented, circular, or irregular s clerotium-like bodies, 65-163 μm diam. | Punctate, olivaceous to black, regular or irregular with brown, superficial, dimidiate, circular pycnothyria, 19-282 μm diam. | Punctate, visible mycelial mat with dull brown, flattened, circular, or irregular sclerotiumlike bodies, 68-145 μm diam. | Punctate, mycelial mat with dark pigmented pycnothyria, 20-100 μm diam. |
a Fungal strain investigated in this study, b Sources of description [9], c Sources of description [12], d Sources of description [7].
PDA: Potato dextrose agar; SNA: Synthetic nutrient-poor agar; WA: 2% water agar.
The partial sequences of the ITS regions (588 bp), and the LSU (810 bp) and mtSSU (781 bp) were obtained for the isolated strains. The BLAST results of the partial ITS region sequences from the five strains revealed a similarity of 98.7-100% with various P. fructicola strains including Pf001, LNHT1506, and 11157. However, strain revealed 84.7% similarities with P. cerophilus 318-07 and 11151, 87.5% similarities with P. gemmifer GTE9a and UIE11b. In the case of the partial LSU sequence, the similarity was 100% with P. fructicola Pf001, LNHT 1506, and 11157, and below 98.1% and 96.9% with P. gemmifer (GTE9a and UIE11b) and P. cerophilus (318-07 and 11151), respectively. The partial mtSSU sequence of the five isolated strains revealed 99.7-99.9% similarity with P. fructicola Pf001, LNHT1506, and 11157, and below 93.4% with P. gemmifer GTE9a and UIE11b and P. cerophilus 318-07 and 11151. Phylogenetic trees were constructed based on the ITS regions, LSU, and mtSSU sequences using the ML method (Fig. 2). The results showed that the five isolated strains were clustered with previously identified strains of P. fructicola. Therefore, the five strains isolated from the apple orchards in this study were identified as P. fructicola.
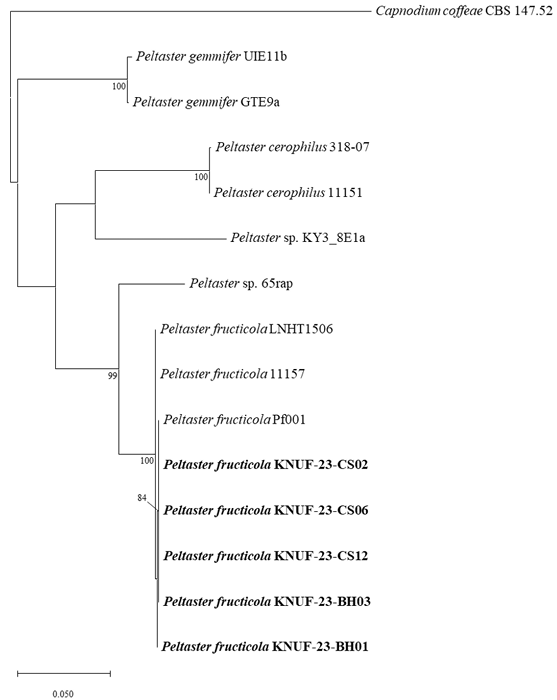
Fig. 2. Maximum likelihood phylogenetic tree of the isolated strains based on the partial sequences of the internal transcribed spacer (ITS) regions, nuclear large subunit (LSU), and mitochondrial small subunit (mtSSU), exhibiting the relationship between Peltaster fructicola with the closest Peltaster species. Capnodium coffeae CBS 147.52 was used as an outgroup. The numbers above the branches indicate bootstrap values (>80%) obtained from 1,000 replicates. The strains isolated in this study are indicated in bold. Bar=0.050 substitutions per nucleotide position.
Punctate mycelial types were observed on the apples’ surface three weeks post-inoculation, with dark, circular, or irregular sclerotium-like bodies and mycelial mats (Fig. 1K-M). The symptoms of SBFS were not observed on the control fruits. The isolated fungal agents were shown to have the same morphological and cultural characteristics as KNUF-23-BH03.
Fungi causing SBFS exhibit a series of mycelial types from only sclerotium-like bodies to mycelial mats without sclerotium-like bodies [4]; some species are distributed worldwide, while others are distributed subcontinentally or locally [6]. The P. fructicola has been previously isolated from crabapple and hawthorn fruits in China and also reported in various regions [12], such as the USA, Norway, and Turkey. In contrast, P. cerophilus and P. gemmifer have only been reported regionally [6]. In Korea, only Gloeodes pomigena and Schizothyrium pomi have been reported as causal agents of SBFS on apples; Peltaster species have not been reported previously [11]. Based on cultural, morphologic, and molecular phylogenic analyses of the strain KNUF-23-BH03 isolated from apples, were identified as P. fructicola that has been reported as the causative agent of SBFS. To our knowledge, this is the first report of the previously unreported species P. fructicola in Korea, as well as the previously unreported disease SBFS on apple fruits caused by P. fructicola in Korea.
The authors declare that they have no potential conflicts of interest.
This research was carried out with the support of “Cooperative Research Program for Agriculture Science and Technology Development (Project No. RS-2021-RD010123)” Rural Development Administration, Republic of Korea.
1. Korea Statistical Information Service. Fruits production [Internet]. Daejeon: Statistics Korea; 2023 [cited 2024 May 15]. Available from https://kosis.kr.
2. Khodadadi F, González JB, Martin PL, Giroux E, Bilodeau GJ, Peter KA, Doyle VP, Aćimović SG. Identification and characterization of Colletotrichum species causing apple bitter rot in New York and description of C. noveboracense sp. nov. Sci Rep 2020;10:1-19. [DOI]
3. Miñarro M, Blázquez MD, Muñoz-Serrano A, Dapena E. Susceptibility of cider apple cultivars to the sooty blotch and flyspeck complex in Spain. Eur J Plant Pathol 2013;135:201-9. [DOI]
4. Gleason ML, Batzer JC, Sun G, Zhang R, Arias MMD, Sutton TB, Crous PW, Ivanović M, McManus PS, Cooley DR, et al. A new view of sooty blotch and flyspeck. Plant Dis 2011;95:368-83. [DOI]
5. Mirzwa-Mróz E, Dzięcioł R, Pitera E, Jurkowski A. Influence of sooty blotch and flyspeck (SBFS) fungi on apple fruits during storage. Acta Sci Pol Hortorum Cultus 2012;11:39-46.
6. Gleason ML, Zhang R, Batzer JC, Sun G. Stealth pathogens: The sooty blotch and flyspeck fungal complex. Annu Rev Phytopathol 2019;57:135-64. [DOI]
7. Rosli H, Batzer JC, Harrington TC, Gleason ML. Peltaster gemmifer: A new species in the sooty blotch and flyspeck species complex from the United States. Mycologia 2018;110:82234. [DOI]
8. Johnson EM, Sutton TB, Hodges CS. Peltaster fructicola: A new species in the complex of fungi causing apple sooty blotch disease. Mycologia 1996;88:114-20. [DOI]
9. Williamson SM, Hodges CS, Sutton TB. Re-examination of Peltaster fructicola, a member of he apple sooty blotch complex. Mycologia 2004;94:885-90. [DOI]
10. Medjedović A, Frank J, Schroers HJ, Oertel B, Batzer JC. Peltaster cerophilus is a new species of the apple sooty blotch complex from Europe. Mycologia 2014;106:525-36. [DOI]
11. Yoon SH, Hong SB, Choi YJ, Lee DH, Lee SY, Choi HY, Lee SH, Choi IS, Kim DG, Kim YH, et al. List of plant diseases in Korea 6th edition. Seoul: The Korean Society of Plant Pathology; 2023. pp. 420-1.
12. Chen C, Gao L, Qu M, Wei X, Li W, Zhang R, Sun G, Gleason ML. Peltaster fructicola, a newly recorded species from China associated with sooty blotch and flyspeck. Mycotaxon 2013;123:265-70. [DOI]
13. Gardes M, Bruns TD. ITS primers with enhanced specificity for Basidiomycetes – application to the identification of mycorrhizae and rusts. Mol Ecol 1993;2:113-8. [DOI]
14. White T, Bruns T, Lee S, Taylor JW. Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics. In: Innis MA, Gelfand DH, Sninsky JJ, White TJ, editors. PCR protocols: A guide to methods and applications. New York: Academic Press, Inc.; 1990. pp. 315-22. [DOI]
15. Rehenr SA, Samuels GJ. Taxonomy and phylogeny of Gliocladium analyzed from nuclear large subunit ribosomal DNA sequences. Mycol Res 1994;98:625-34. [DOI]
16. Vilgalys R, Hester M. Rapid genetic identification and mapping of enzymatically amplified ribosomal DNA from several Cryptococcus species. J Bacteriol 1990;172:4238-46. [DOI]
17. Zoller S, Scheidegger C, Sperisen C. PCR primers for the amplification of mitochondrial small subunit ribosomal DNA of lichen-forming Ascomycetes. Lichenologist 1999;31:511-6. [DOI]
18. Tamura K, Nei M. Estimation of the number of nucleotide substitutions in the control region of mitochondrial DNA in humans and chimpanzees. Mol Biol Evol 1993;10:512-26.
19. Tamura K, Stecher G, Kumar S. MEGA11: Molecular evolutionary genetics analysis version 11. Mol Biol Evol 2021;38:3022-7. [DOI]
20. Johnson EM, Sutton TB, Hodges CS. Etiology of apple sooty blotch disease in North Carolina. Phytopathology 1997;87:88-95. [DOI]
21. Frank J, Crous PW, Groenewald JZ, Oertel B, Hyde KD, Phengsintham P, Schroers HJ. Microcyclospora and Microcyclosporella: novel genera accommodating epiphytic fungi causing sooty blotch on apple. Persoonia 2010;24:93-105. [DOI]
22. Jung B, Kim S, Lee J. Microcyle conidiation in filamentous fungi. Mycobiology 2014;42:1-5. [DOI]