Tae-Gyeong Kim1, Leonid N. Ten2, Soo-Min Hong1, Seong-Keun Lim1, Seung-Yeol Lee1,2, and HeeYoung Jung1,2,*
1College of Agriculture and Life Sciences, Kyungpook National University, Daegu 41566, Korea
2Institute of Plant Medicine, Kyungpook National University, Daegu 41566, Korea
*Correspondence to heeyoung@knu.ac.kr
Korean Journal of Mycology (Kor J Mycol) 2024 September, Volume 52, Issue 3, pages 155-163.
https://doi.org/10.4489/kjm.520301
Received on June 09, 2024, Revised on July 10, 2024, Accepted on August 09, 2024, Published on Sep 30, 2024.
Copyright © The Korean Society of Mycology.
This is an Open Access article which is freely available under the Creative Commons Attribution-NonCommercial 4.0 International License (CC BY-NC) (https://creativecommons.org/licenses/by-nc/4.0/).
A fungal strain designated KNUF-22-016 was isolated from a soil sample collected in Cheoltan Mountain in Yeongju, Korea. The cultural and morphological characteristics of this strain closely resembled those of Hamigera ingelheimensis. The colonies formed by the isolate appeared light orange to dark orange on Czapek yeast extract agar, its conidiophores were rarely branched, smooth-walled, measuring 109.9‒864.8 × 4.1‒7.4 µm, with its smoothwalled conidia being 2.3‒4.6 × 1.9‒3.5 µm in size. Phylogenetic analysis using concatenated sequences of the internal transcribed spacer (ITS) regions and the Mcm7, RPB2 and Tsr1 genes confirmed the affiliation of strain KNUF-22-016 with H. ingelheimensis. To the best of our knowledge, this fungus has not been previously documented in Korea.
Hamigera, Multilocus sequence analysis, Soil-inhabiting fungi, Unreported fungi
The genus Hamigera was first established to accommodate two species that were formerly classified under the genus Talaromyces (T. avellaneus and T. striatus). These two species could be distinguished from other members of the genus because they formed individual asci rather than in chains. This distinctive trait led Stolk and Samson in 1971 to propose the genus Hamigera through the reclassification of T. avellaneum and T. striatus as Hamigera avellanea (the type species) and H. striata, respectively [1]. Since then, the taxonomic classification of H. striata has changed a few times and is currently recognized as Pseudohamigera striata [2]. In 2010, the taxonomic affiliations of several fungal isolates from the Agricultural Research Service Culture Collection (NRRL) were examined, and a multilocus sequence analysis (MLSA) using the Mcm7, RPB2, and Tsr1 genes was conducted [3]. From the analyzed isolates, six strains were identified as new species within the genus Hamigera, namely H. fusca, H. inflata, H. insecticola, H. pallida, H. paravellanea, and H. terricola [3]. Another member of the genus Hamigera, H. ingelheimensis, underwent a series of reclassifications prior to being assigned its current taxonomic affiliation. Initially, it was classified as Penicillium ingelheimensis, after which it was reclassified as Merimbla ingelheimensis based on a phylogenetic analysis distinguishing it from closely related species such as H. avellanea [4]. Following the Melbourne Code [5], which allows only a single name for a fungus, and to reflect the phylogenetic position of the species, M. ingelheimensis was later reclassified as H. ingelheimensis [6]. Recently, Houbraken et al. (2020) proposed the reclassification of Talaromyces brevicompactus to Hamigera brevicompacta based on phylogenetic analysis using concatenated sequences of the BenA, CaM, and RPB2 genes [2]. As a result, the genus Hamigera currently encompasses nine species: H. avellanea, H. fusca, H. inflata, H. ingelheimensis, H. insecticola, H. pallida, H. paravellanea, H. terricola, and H. brevicompacta.
This study sought to identify previously unreported Hamigera species in Korea, isolated from a soil sample collected in Cheoltan mountain, Yeongju. This investigation was conducted as part of our research initiative focused on discovering indigenous Korean fungal species. The isolated strain KNUF-22-016 was characterized through both morphological and molecular analyses, and its taxonomic placement within the genus was confirmed utilizing an MLSA approach.
In 2023, soil samples were collected from Cheoltan Mountain in Yeongju, Gyeongsangbuk-do, Korea (36° 49’52.3”N, 128°37’46.1”E) and transported to the laboratory for further analysis. Fungi were isolated using the dilution plating method. Each soil sample was mixed with 10 mL of sterile distilled water, vortexed, serially diluted, and spread onto potato dextrose agar (PDA; Difco, Detroit, MI, USA) plates. The PDA plates were then incubated at 25℃ for one week. Afterward, single fungal colonies were transferred to new plates and incubated under the same conditions. Several fungal strains were isolated and preliminarily identified by sequencing the internal transcribed spacer (ITS) regions. Among them, strain KNUF-22-016 was recognized as a fungal species that had not been previously identified in Korea. Therefore, this strain was selected for comprehensive analysis, including morphological and molecular phylogenetic analyses. Stock culture of the strain, namely, KNUF-22-016 (NIBRFGC000509833) was deposited in the National Institute of Biological Resources (NIBR) as metabolically inactive cultures.
After one week of incubation at 25℃ on Czapek yeast extract agar (CYA; MBcell, Seoul, Korea), malt extract agar (MEA; Difco, Detroit, MI, USA), and glycerol nitrate agar (G25N), the morphological features of isolate KNUF-22-016, including color, shape, and size, were recorded [3]. Additionally, cultures on CYA were incubated at 5℃ and 37℃ for one week [3]. The fungal characteristics were examined using a light microscope (BX-50, Olympus, Tokyo, Japan).
Genomic DNA was extracted from the mycelia using the HiGene Genomic DNA Prep Kit (Biofact, Daejeon, Korea) according to the manufacturer’s instructions. For molecular analysis, the ITS regions, minichromosome maintenance complex component 7 (Mcm7), the second largest subunit of RNA polymerase II (RPB2), and ribosome biogenesis protein (Tsr1) genes were amplified via PCR [3]. Specifically, the ITS regions, the partial Mcm7 gene, the partial RPB2 gene, and the partial Tsr1 gene were respectively amplified using primers ITS1F/ITS4 [7,8], 709F/1447R [9], 5F/7CR [10], and 1459F/2308R [9]. Subsequently, the sequences of the amplified ITS regions, as well as those of the Mcm7, RPB2, and Tsr1 genes, were deposited in the GenBank database under accession numbers LC799815, LC800019, LC800020, and LC800021, respectively.
The sequences of our isolates were compared with reference sequences from the GenBank database of the National Center for Biotechnology Information (NCBI) using the Basic Local Alignment Search Tool (BLAST). The sequences of the ITS regions, the Mcm7, RPB2, and Tsr1 genes were concatenated and phylogenetic trees were constructed using the maximum likelihood (ML), neighbor-joining (NJ), and maximum parsimony (MP) algorithms based on the Kimura model [11]. Phylogenetic analysis was conducted using the MEGA 7 (https://www.megasoftware.net/) software utilizing bootstrap values from 1,000 replications [12]. The reference sequences from the NCBI GenBank database are listed in Table 1.
Table 1. List of species used in phylogenetic analysis along with their GenBank accession numbers.
| Species | Strain | GenBank accession numbers | |||
|---|---|---|---|---|---|
| ITS | Mcm7 | RPB2 | Tsr1 | ||
| Hamigera avellanea | NRRL 1938T | AF454075 | GU092852 | EU021627 | GU092726 |
| Hamigera avellanea | NRRL 58017 | GU092954 | GU092896 | GU092917 | GU092727 |
| Hamigera brevicompacta | CBS 102661T | NR_160208 | – | MN969203 – | |
| Hamigera fusca | NRRL 35721 | GU092939 | GU092888 | GU111760 | GU092717 |
| Hamigera fusca | NRRL 35601T | NR_137734 | GU092879 | GU111755 | GU092715 |
| Hamigera inflata | NRRL 58014T | NR_137736 | GU092895 | GU092908 | GU092725 |
| Hamigera ingelheimensis | NRRL 3522 | GU092960 | GU092871 | GU092911 | GU092730 |
| Hamigera ingelheimensis | NRRL 2110T | MH856108 | GU092856 | GU092912 | GU092728 |
| Hamigera ingelheimensis | KNUF-22-016 | LC799815 | LC800019 | LC800020 | LC800021 |
| Hamigera insecticola | NRRL 35386T | NR_137684 | GU092872 | GU111754 | GU092718 |
| Hamigera insecticola | NRRL 35442 | EF634416 | GU092873 | GU092907 | GU092722 |
| Hamigera pallida | NRRL 35718T | NR_137737 | GU092885 | GU111758 | GU092710 |
| Hamigera paravellanea | NRRL 35714 | GU092953 | GU092882 | GU092918 | GU092737 |
| Hamigera paravellanea | NRRL 35720T | NR_137738 | GU092887 | GU092919 | GU092738 |
| Hamigera terricola | NRRL 29055T | NR_137735 | GU092860 | GU111751 | GU092712 |
| Hamigera terricola | NRRL 35717 | GU092945 | GU092884 | GU111757 | GU092708 |
| Hamigera brevicompacta | CBS 102661T | NR_160208 | – | MN969203 – | |
| Pseudohamigera striata | NRRL 717T | NR_145139 | GU092901 | GU092928 | GU092697 |
ITS: internal transcribed spacer regions; Mcm7: minichromosome maintenance complex component 7; RPB2: the second largest subunit of RNA polymerase II;
Tsr1: Tsr1 ribosome maturation factor. TType strain. The strain isolated in this study is indicated in boldface.
After 7 days of incubation at 25℃ on CYA, the colony size of strain KNUF-22-016 reached 67.1‒69.8 mm. The colony exhibited a variety of colors ranging from light orange to dark orange, with a pale pink reverse side (Fig. 1A and 1B). In contrast, at 37℃, colony size decreased to 37.8‒39.1 mm, indicating a suboptimal temperature for growth (Fig. 1C and 1D). After 7 days of incubation at 25℃ on MEA, the colonies measured 61.2–62.9 mm, displaying a light pinkish cinnamon color with a pale pinkish reverse side (Fig. 1E and 1F). No growth was observed on G25N medium under the same culture conditions or on CYA after 7 days of incubation at 5℃. Conidiophores, arising from the colony surface, ranged in size from 109.9‒864.8 × 4.1‒7.4 µm (Fig. 1G and 1H). Apical whorls of metulae typically varied in size from 4.6‒14.2 × 2.4‒5.7 µm, each bearing five or more phialides measuring 6.5‒10.6 × 1.6‒3.3 µm (Fig. 1G and 1H). The conidia were observed to be 2.3‒4.6 × 1.9‒3.5 µm in size, with smooth walls (Fig. 1I). Based on both its cultural and morphological traits, the isolated strain KNUF-22-016 appears to be closely affiliated with H. ingelheimensis [3]. In contrast, KNUF-22-016 differed from its close relative, H. avellanea, in several aspects, including colony color, colony size (61.2‒62.9 mm vs. 45‒70 mm on MEA), conidiophore size (109.9‒864.8 × 4.1‒7.4 µm vs. 200‒500 × 3‒5 µm), and larger metulae (4.6‒14.2 × 2.4‒5.7 µm vs. 5‒12 × 3‒5 µm) (Table 2).
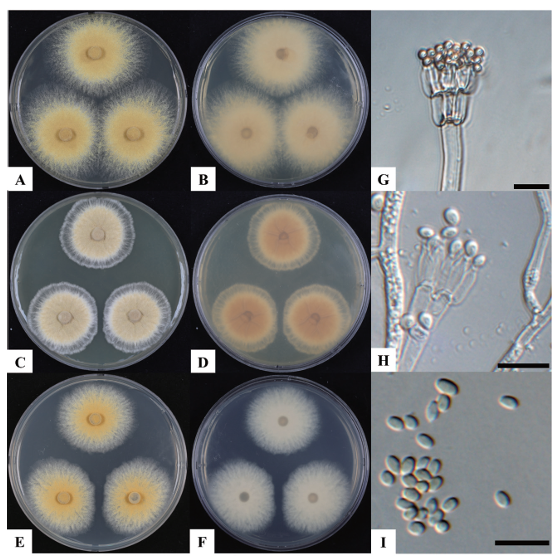
Fig. 1. Cultural and morphological characteristics of Hamigera ingelheimensis KNUF-22-016. A, B: colony on Czapek yeast extract agar (CYA) after 7 days at 25℃; C, D: colony on CYA after 7 days at 37℃; E, F: colony on malt extract agar (MEA) after 7 days at 25℃; G, H: conidiophores; I: conidia. Scale bars G-I = 10 µm.
Table 2. Comparison of morphological characteristics of KNUF-22-016 with reference species Hamigera ingelheimensis and H. avellanea.
| Characteristics | Hamigera ingelheimensis KNUF-22-016a | H. ingelheimensisb | H. avellaneac | |
|---|---|---|---|---|
| Colony | Color | CYA: light orange to dark orange and pink in reverse; MEA: light pinkish cinnamon | CYA: light salmon orange to capucine orange, pale pink near safrano pink in reverse; MEA: light pinkish cinnamon to cinnamon buff | CYA: conidial areas pinkish, deep red shades near Indian red in reverse; MEA: pale yellow to pinkish, vinaceous purple in reverse; G25N: light buff in reverse |
| Size (diam.) | CYA: 67.1‒69.8 mm; MEA: 61.2‒62.9 mm in 7 days at 25℃ | CYA: 55‒65 mm; MEA: 70 mm in 7 days at 25℃ | CYA: 50‒70 mm; MEA: 45‒70 mm; G25N: 23‒24 mm in 7 days at 25℃ | |
| Shape | CYA: plane, velutinous, no exudate; MEA: thin, plane, velutinous, low, heavy sporulation | CYA: plane, velutinous, no exudate; MEA: thin, plane, velutinous, low, heavy sporulation | CYA: lanose to velvety, low; MEA: thin, low, sporulating well; G25N: plane, low, thin | |
| Conidiophores | Size (μm) | 109.9‒864.8 × 4.1‒7.4 | 100‒800 × 4‒7 | 200‒500 × 3‒5 |
| Shape | rarely branched, smooth walls, arising from colony surface | rarely branched, smooth walls, arising from colony surface | rarely branched, arising from colony surface, smooth walls | |
| Metulae | Size (μm) | 4.6‒14.2 × 2.4‒5.7 | 3‒14 × 3‒6 | 5‒12 × 3‒5 |
| Conidia | Size (μm) | 2.3‒4.6 × 1.9‒3.5 | 3.5‒5.0 × 2‒3 | 3‒5 × 2‒3 |
| Shape | smooth walls | smooth walls | smooth walls | |
| Philaides | Size (μm) | 6.5‒10.6 × 1.6‒3.3 | 7‒9 × 2‒3 | 5‒9 × 2.5‒3.5 |
| Shape | acerose to ampulliform | acerose to ampulliform | ampulliform |
CYA: Czapek yeast extract agar; MEA: malt extract agar; G25N: glycerol nitrate agar; Diam.: diameter.
aFungal strain used in this study; bSource of description [3]; cSources of description [1,3].
The amplicons obtained from the ITS, Mcm7, RPB2, and Tsr1 loci of strain KNUF-22-016 were 591, 681, 1020, and 781 bp long, respectively. The ITS sequence of the isolate exhibited 99.6‒100% identity with several strains of H. ingelheimensis, including NRRL 29059 (GU092961), NRRL 3522 (GU092960), and CBS:163.42 (MH856108). Additionally, strain KNUF-22-016 exhibited a close relationship with H. avellanea NRRL 1938 (98.9% similarity, AF454075), H. paravellanea NRRL 35720 (98.9%, NR_137738), H. brevicompacta CBS 102661 (98.7%, NR_160208), and H. fusca NRRL 35601 (98.6%, NR_137734). Based on the similarity of the Mcm7 gene sequence, H. ingelheimensis NRRL 2110 (100% similarity, GU092856), H. paravellanea NRRL 35720 (98.5%, GU092887), H. avellanea NRRL 58017 (98.3%, GU092896), and H. inf lata NRRL 58014 (97.3%, GU092895) were the closest phylogenetic relatives of strain KNUF-22-016. The partial RPB2 gene sequence of the isolate was 100, 99.2, 99.0, 98.9, and 97.4% similar to those of H. ingelheimensis NRRL 58707 (GU092909), H. avellanea NRRL 1938 (EU021627), H. paravellanea NRRL 35720 (GU092919), H. brevicompacta CBS 102661 (MN969203), and H. terricola NRRL 58014 (GU092908), respectively. The Tsr1 sequence of strain KNUF-22-016 shared 100, 98.7, and 97.9% identity with H. ingelheimensis NRRL 2110 (GU092728), H. avellanea NRRL 1938 (GU092726), and H. paravellanea NRRL 35714 (GU092737), respectively. These findings demonstrated that several Hamigera species are closely related to strain KNUF-22-016. Although the isolate exhibited 100% similarity with various strains of H. ingelheimensis based on the sequences of the four molecular markers examined herein, it is evident that the strain could have not been precisely identified based on only a single molecular marker. Therefore, multilocus sequence analysis was conducted using concatenated sequences of the ITS regions, as well as the Mcm7, RPB2, and Tsr1 genes. This approach successfully allowed for the identification of the six members of the genus Hamigera from the current nine validated species [3]. The ML phylogenetic tree based on the concatenated sequences clearly demonstrated that the phylogenetic characteristics of KNUF-22-016 were consistent with those of H. ingelheimensis (Fig. 2). The same topology of the phylogenetic tree was also obtained using the NJ and MP algorithms, as indicated by the filled circles in Fig. 2, further supporting the affiliation of the isolate. Unfortunately, the sequences of the Mcm7 and Tsr1 genes for the recently described H. brevicompacta were unavailable in the GenBank database. However, the topology of the phylogenetic tree constructed using the concatenated sequences of only ITS regions and the RPB2 gene (tree not shown) was similar to that of the abovementioned phylogenetic tree, confirming the differentiation of KNUF-22-016 from H. brevicompacta at the species level. The results of both morphological and phylogenetic analyses revealed that KNUF-22-016 is a strain belonging to H. ingelheimensis. To the best of our knowledge, this study represents the first report of this fungal species in Korea.
Various biologically active compounds have been identified in members of the genus Hamigera. Among the nine species mentioned above, the production of secondary metabolites has been more extensively studied in H. avellanea. The antifungal compounds hamigerone, dihydrohamigerone, and avellaneanone, the potential antimalarial compounds hamavellone B and hamigeromycins B, the bioactive alkaloid pseurotin A, and the anthraquinone pigments emodin, ω-hydroxyemodin, and emodic acid have been isolated from BCC 17816, KUFA 0732, and other strains of H. avellanea [13-16]. Among these, pseurotin A is known for its cytotoxic activity against mouse leukemia P388, human leukemia HL60, human lung carcinoma A-549, and human hepatic carcinoma BEL-7402 cell lines, as well as for its inhibition of the fungal chitin synthase and anti-inflammatory activity [17]. Emodin possesses a wide range of bioactive properties, including antibacterial, anticancer, cardioprotective, antioxidant, anti-fibrotic, and antiinflammatory effects [18]. Eight species of the genus Hamigera, excluding H. ingelheimensis, produced avellanins A and B as the most common metabolites, exhibiting inhibitory activity against apolipoprotein B production [19]. Interestingly, H. ingelheimensis produced other structurally related compounds, including avellanin C and the cyclic hexapeptide PF1171C [20]. Avellanin C is known for its quorum-sensing (QS) inhibiting activity against Staphylococcus aureus [20]. The inhibition of QS in S. aureus can reduce bacterial virulence, thereby enhancing the host’s innate immune response and limiting inflammation [21]. In contrast to H. avellanea, the production of bioactive compounds by H. ingelheimensis has not been thoroughly investigated. Therefore, isolate KNUF-22-016 can be considered a valuable strain for further study of this species in Korea.
This study provides the first record of the occurrence of H. ingelheimensis in Korea, contributing to the discovery of indigenous Korean fungal species. Additional research is needed to gain more comprehensive insights into the geographic distribution of H. ingelheimensis and its ecological and biological roles in Korean ecosystems, as well as to explore its potential therapeutic properties.
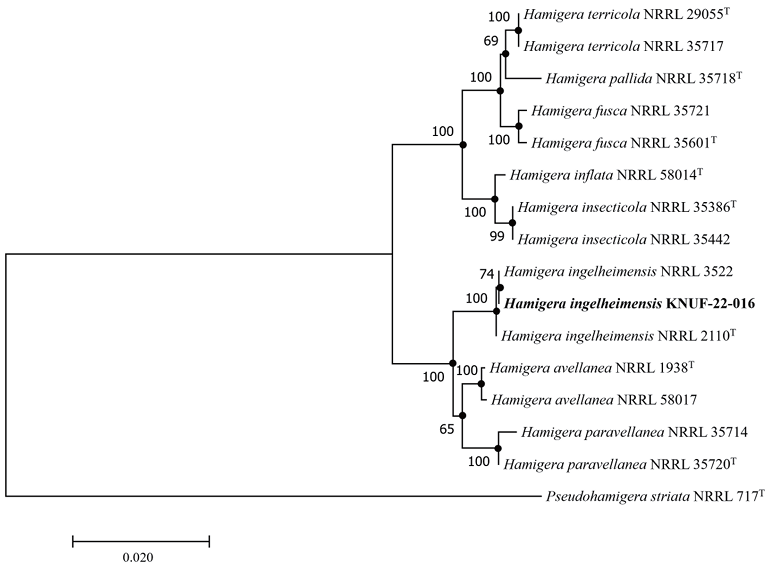
Fig. 2. Maximum-likelihood phylogenetic tree based on the combined sequences of internal transcribed spacer (ITS) regions, minichromosome maintenance complex component 7 (Mcm7), the second largest subunit of RNA polymerase II (RPB2), and Tsr1 ribosome maturation factor (Tsr1) showing the phylogenetic position of strain KNUF-22-016 among Hamigera species. Bootstrap values greater than 60% (based on 1,000 replications) are shown at branch points. The filled circles indicate that the corresponding nodes were also recovered in trees generated using the neighbor-joining and maximum parsimony algorithms. The isolated strain is indicated in bold. Pseudohamigera striata NRRL 717T was used as an outgroup. Bar, 0.02 substitutions per nucleotide position.
The authors declare that they have no potential conflicts of interest.
This study was supported by the National Institute of Biological Resources, funded by the Ministry of Environment of the Republic of Korea [NIBR202203112].
1. Stolk AC, Samson RA. Studies on Talaromyces and related genera I. Hamigera gen. nov. and Byssochalmys. Persoonia 1971;6:341-57.
2. Houbraken J, Kocsube S, Visagie CM, Yilmaz N, Wang XC, Meijer M, Kraak B, Hubka V, Besnch K, Samson RA, et al. Classification of Aspergillus, Penicillium, Talaromyces and related genera (Eurotiales): An overview of families, genera, subgenera, sections, series, and species. Stud Mycol 2020;165-9. doi: 10.1016/j.simyco.2020.05.002 [DOI]
3. Peterson SW, Jurjevic Z, Bills GF, Stchigel AM, Vega FE. Genus Hamigera, six new species and multilocus DNA sequence-based phylogeny. Mycologia 2010;102:847-64. doi: 10.3852/09-268 [DOI]
4. Pitt JI. The genus Penicillium and its teleomorphic states Eupenicillium and Talaromyces. London: Academic Press; 1979.
5. Mcneill J, Barrie FR, Buck WR, Demoulin V, Greuter W, Hawksworth DL, Herendeen PS, Knapp MSK, Prado J, Prud’homme Van Reine WF. International code of nomenclature for algae, fungi, and plants (Melbourne code). Regnum Vegetabile 154. Koenigstein: Koeltz Scientific Books; 2012.
6. Igarashi Y, Hanafusa T, Gohda F, Peterson S, Bills G. Species-level assessment of secondary metabolite diversity among Hamigera species and a taxonomic note on the genus. Mycologia 2014;5:102-9. doi: 10.1080/21501203.2014.917736 [DOI]
7. Gardes M, Bruns TD. ITS primers with enhanced specificity for basidiomycetes-application to the identification of mycorrhizae and rusts. Mol Ecol 1993;2:113-8. doi: 10.1111/j.1365294X.1993.tb00005.x [DOI]
8. White TJ, Bruns T, Lee S, Taylor J. Amplification, and direct sequencing of fungal ribosomal RNA genes for phylogenetics. PCR protocols: a guide to methods and applications. In: Innis MA, Gelfand DH, Sninsky JJ, White TJ, editors. New York AP 1990;18:315-22. [DOI]
9. Schmitt I, Crespo A, Divakar PK, Fankhauser JD, HermanSackett E, Kalb K, Nelsen MP, Nelson NA, Rivas-Plata E, Shimp AD, Widhelm T, Lumbsch HT. New primers for promising single copy genes in fungal phylogenetics and systematics. Persoonia 2009;23:35-40. doi: 10.3767/003158509X470602 [DOI]
10. Peterson SW. Phylogenetic analysis of Aspergillus species using DNA sequences from four loci. Mycologia 2008;100:205-26. doi: 10.1080/15572536.2008.11832477 [DOI]
11. Kimura M. A simple method for estimating evolutionary rates of base substitutions through comparative studies of nucleotide sequences. J Mol Evol 1980;16:111-20. [DOI]
12. Kumar S, Stecher G, Tamura K. MEGA7: Molecular Evolutionary Genetics Analysis Version 7.0 for Bigger Datasets. Mol Biol Evol 2016;33:1870-4. doi: 10.1093/molbev/msw054 [DOI]
13. Breinholt J, Kjaer A, Olsen CE, Rassing BR, Rosendahl CN. Hamigerone and dihydrohamigerone: two acetate-derived, antifungal metabolites from Hamigera avellanea. Acta Chem Scand 1997;51:1241-4. [DOI]
14. Isaka M, Chinthanom P, Veeranondha S, Supothina S, Luangsa-ard JJ. Novel cyclopropyl diketones and 14-membered macrolides from the soil fungus Hamigera avellanea BCC 17816. Tetrahedron 2008;64:11028-33. doi: 10.1016/j.tet.2008.09.077 [DOI]
15. Isaka M, Chinthanom P, Kongthong, S, Supothina S, Ittiworapong S. Hamigeromycins CG, 14-membered macrolides from the fungus Hamigera avellanea BCC 17816. Tetrahedron 2010;66:955-61. doi: 10.1016/j.tet.2009.11.101 [DOI]
16. Klaram R, Dethoup T, Machado FP, Gales L, Kumla D, Hafez Ghoran S, Sousa E, Mistry S, Silva AMS, Kijjoa A. Pentaketides and 5-p-hydroxyphenyl-2-pyridone derivative from the culture extract of a marine sponge-associated fungus Hamigera avellanea KUFA0732. Mar Drugs 2023;21:344. doi: 10.3390/md21060344 [DOI]
17. Abdelwahed KS, Siddique AB, Mohyeldin MM, Qusa MH, Goda AA, Singh SS, Ayoub NM, King JA, Jois SD, El Sayed KA. Pseurotin A as a novel suppressor of hormone dependent breast cancer progression and recurrence by inhibiting PCSK9 secretion and interaction with LDL receptor. Pharmacol Res. 2020;158:104847. doi: 10.1016/j.phrs.2020.104847 [DOI]
18. Luo N, Fang J, Wei L, Sahebkar A, Little PJ, Xu S, Luo C, Li G. Emodin in atherosclerosis prevention: pharmacological actions and therapeutic potential. Eur J Pharmacol. 2021;890:173617. doi: 10.1016/j.ejphar.2020.173617 [DOI]
19. Igarashi Y, Hanafusa T, Gohda F, Peterson S, Bills G. Species-level assessment of secondary metabolite diversity among Hamigera species and a taxonomic note on the genus, Mycology. 2014;5:102-9. doi: 10.1080/21501203.2014.917736 [DOI]
20. Igarashi Y, Gohda F, Kadoshima T, Fukuda T, Hanafusa T, Shojima A, Nakayama J, Bills G, Peterson S. Avellanin C, an inhibitor of quorumsensing signaling in Staphylococcus aureus, from Hamigera ingelheimensis. J Antibiot. 2015;68:707-10. doi: 10.1038/ja.2015.50 [DOI]
21. Daly SM, Elmore BO, Kavanaugh JS, Triplett KD, Figueroa M, Raja HA, El-Elimat T, Crosby HA, Femling JK, Cech NB, Horswill AR, Oberlies NH, Hall PR. Omega-Hydroxyemodin limits Staphylococcus aureus quorum sensing-mediated pathogenesis and inflammation. Antimicrob Agents Chemother 2015;59:2223-35. doi: 10.1128/AAC.04564-14 [DOI]