Mohammad Hamizan Azmi1, Seong-Keun Lim1, Sang-Min Lee1, Dae-Hoon Kang1, Jin-Sil Choi1, Gwang-Jae Lim1, Jun-Woo Choi1, Min-Kyu Kim1, Seung-Yeol Lee1,2,*, and Hee-Young Jung1,2
1Department of Plant Medicine, Kyungpook National University, Daegu 41566, Korea
2Institute of Plant Medicine, Kyungpook National University, Daegu 41566, Korea
*Correspondence to leesy1123@knu.ac.kr
Korean Journal of Mycology (Kor J Mycol) 2024 September, Volume 52, Issue 3, pages 165-175.
https://doi.org/10.4489/kjm.520302
Received on July 16, 2024, Revised on August 20, 2024, Accepted on August 20, 2024, Published on Sep 30, 2024.
Copyright © The Korean Society of Mycology.
This is an Open Access article which is freely available under the Creative Commons Attribution-NonCommercial 4.0 International License (CC BY-NC) (https://creativecommons.org/licenses/by-nc/4.0/).
During an exploration of fungal diversity in Korean soil, this study isolated two fungal strains, designated KNUF-23-321L and KNUF-23-314, and identified them as previously unrecorded species. Morphological and cultural features were examined for initial classification. The characteristics of strains KNUF-23-321L and KNUF-23-314 matched those of Cladophialophora floridana SR3028T, and Chloridium setosum CGMCC 3.20741T, respectively. To further pinpoint their identities and evolutionary relationships, molecular phylogenetic analyses were conducted using the internal transcribed spacer (ITS) regions and 28S rDNA large subunit (LSU) on strain KNUF-23-321L which showed high similarity of 98.6 and 100%, respectively, with strain C. floridana SR3028T. For the strain KNUF-23-314, it showed a high similarity of 100% with strain Ch. setosum CBS 263.76A with both ITS regions and LSU genes. These combined morphological and phylogenetical analyses revealed that KNUF-23-321L was identified as C. floridana, and KNUF-23-314 was identified as Ch. setosum. To the best of our knowledge, these are the first reports of C. floridana and Ch. setosum in Korea.
Chloridium setosum, Cladophialophora floridana, phylogeny, soil fungi, unreported species
Soil, as the primary habitat within natural ecosystems, harbors a diverse array of microorganisms [1]. Among the diverse soil microorganisms, fungi can have positive effects on plants and soils as mycorrhizal fungi, decomposers, and biosorbents [2]. It is estimated that more than 1.5 million species of fungi exist worldwide, with over 150,000 described species reported to date [3].
The fungal genus Cladophialophora, of the phylum Ascomycota, class Eurotiomycetes, order Chaetothyriales, and family Herpotrichiellaceae, is morphologically characterized by conidia that germinate through blastic, acropetal conidiogenesis, forming interconnected chains of conidia [4,5]. This genus is widely distributed in plant residues, soil, skin lesions, and lower extremities and has been reported to be pathogenic in plants and mammals, including humans, with three species affecting plants and 11 humans [5,6]. It was first described in the 1980s, and currently, 68 species are registered worldwide [5]. Three species, C. hostae, C. pucciniophila, and C. lanosa, have been reported in Korea, which were isolated from rust fungi conidia, daylily (Hemerocallis sp.), and soil, respectively [7-9].
The genus Chloridium belongs to the phylum Ascomycota, class Sordariomycetes, order Chaetosphaeriales, and family Chaetosphaeriaceae [10]. Chloridium species have been isolated from a variety of sources, including soil, trees, and mosses, and are known to be endophytic or saprophytic in association with a variety of plants, including grasses and trees [11]. The first reported Chloridium species was Ch. virescens, and a total of 38 species have been identified to date [10]. However, only two, Ch. virescens and Ch. apiculatum, have been isolated and reported in Korea, and domestic studies of this genus are relatively scarce compared to overseas studies.
To expand the known fungal species diversity in Korea and discover new species, fungi were isolated from a soil specimen obtained in Korea. In this study, two fungal strains, designated as KNUF-23-321L and KNUF-23-314, were subjected to a comprehensive analysis assessing their cultural, molecular, and morphological attributes. The findings are documented and reported below.
The fungal strains used in this study were isolated from the soil samples collected in Gachangmyeon, Daegu (35°47’14.1″N 128°33’19.7″E) and Geochang-gun, Gyeongsangnam-do (35°50’01.8″N, 127°47’44.0″E) in Korea. Using an autoclaved spatula, soil samples were taken from the field to a depth of 15-30 cm. The samples were then air-dried and kept at 4℃ in a plastic bag. For isolation, 1 g from each soil sample was suspended in 10 mL of sterile distilled water, gently vortexed, and serially diluted. A 100 μL volume was then spread onto potato dextrose agar (PDA; Difco, Detroit, MI, USA) plates, which were then cultured for 2-3 days at 25℃ [12]. Germinating single colonies were then transferred to new PDA plates at 25℃, and based on specific characteristics, pure cultures of isolates KNUF-23-314 and KNUF23-321L were selected for further molecular analyses and cultural and morphological characterization. The specimens have been stored at the National Institute of Biological Resources (NIBR) and are identified by the accession numbers NIBRFGC000510718 and NIBRFGC000510719, respectively.
Following previous studies, the cultural and morphological characteristics of the two fungal strains were assessed using cultural media based on their genus. The strain KNUF-23-321L was cultured on PDA for 20 days at 25℃, while KNUF-23-314 was cultured on PDA for 28 days at 25℃ [6]. Additionally, KNUF-23-321L was grown for 28 days at 25℃ on malt extract agar (MEA; Difco, Detroit, MI, USA) [11]. The growth of the fungi was measured, and characteristics of the colonies, such as their color, form, and dimensions, were documented. Morphological features were examined using a light microscope (BX-50; Olympus, Tokyo, Japan).
For the molecular identification of strains KNUF-23-314 and KNUF-23-321L, total genomic DNA was extracted using the HiGene™ Genomic DNA Prep Kit for fungi (Biofact, Daejeon, Korea). The internal transcribed spacer (ITS) region of the total genomic DNA extracted from the samples was amplified using the ITS1F/ITS4 primer pair, and the large subunit (LSU) gene of the 28S rDNA was amplified using the LROR/LR5 primer pair [13-15]. Amplification was confirmed by electrophoresis on 1% agarose gels stained with ethidium bromide. The resulting amplification products were purified utilizing ExoSAP-IT (Thermo Fisher Scientific, Waltham, MA, USA) and then subjected to the sequencing services provided by Solgent (Daejeon, Korea).
The sequences of KNUF-23-314 and KNUF-23-321L were analyzed for similarity against those available in the National Center for Biotechnology Information (NCBI) database utilizing the Basic Local Alignment Search Tool (BLAST), and a number of related sequences were downloaded from the database for phylogenetic analyses (Table 1). Phylogenetic trees were constructed based on concatenated ITS regions and LSU gene sequences using the neighbor-joining (NJ) method in MEGA version X [16,17]. The evolutionary distance matrices for the NJ analysis were produced using Kimura’s two-parameter model, and bootstrap values derived from 1,000 resamples were produced to provide branch support [18].
Table 1. GenBank strain and accession numbers of the strains utilized for the phylogenetic analyses in this study
| Species Name | Strain Number | GenBank accession number | |
|---|---|---|---|
| ITS | LSU | ||
| Chloridium bellum var. bellum | CBS 709.73A | NR_185446 | OP455466 |
| Chloridium bellum var. bellum | CBS 709.73BT | OP455361 | OP455467 |
| Chloridium bellum var. luteum | CBS 141.54T | OP455362 | OP455469 |
| Chloridium caudigerum | CBS 145489 | OP455379 | OP455486 |
| Chloridium caudigerum | CBS 145433 | OP455377 | OP455484 |
| Chloridium guttiferum | CBS 126073 | MH864068 | MH875524 |
| Chloridium jilinense | NN046507 | OL627659 | OL655058 |
| Chloridium setosum | CBS 263.76A | OP455427 | OP455534 |
| Chloridium setosum | KNUF-23-314 | PP727493 | PP727510 |
| Chloridium virescens | CBS 145487 | OP455441 | OP455549 |
| Chloridium virescens | CBS 138683 | OP455431 | OP455539 |
| Cladophialophora boppii | CBS 126.86T | EU103997 | NG_058762 |
| Cladophialophora chaetospira | CBS 491.70 | EU035405 | EU035405 |
| Cladophialophora chaetospira | CBS 514.63 | MH858340 | KF928513 |
| Cladophialophora floridana | KNUF-23-321L | PP727494 | PP727511 |
| Cladophialophora floridana | SR1004 | AB986344 | AB986344 |
| Cladophialophora floridana | SR3028T | AB986343 | AB986343 |
| Cladophialophora multiseptata | FMR 10591T | HG003668 | HG003671 |
| Cladophialophora potulentorum | CBS 112222 | EU03540 | EU03540 |
| Cladophialophora potulentorum | CBS 114772 | EU035410 | EU035410 |
| Cladophialophora tortuosa | BA4b006T | AB986424 | AB986424 |
| Exophiala oligosperma | CBS 725.88T | AY163551 | NG_059201 |
| Menispora ciliata | CBS 984.70 | MH860017 | MH871802 |
ITS: internal transcribed spacer regions; LSU: 28S rDNA large subunit gene.
TType strain.
The strains identified in this study are indicated in bold.
On PDA, the colonies achieved a growth of 32‒34 mm in diameter after 28 days at 25℃, with the colonies appearing olivaceous-black with a dry, velvety, and ovoid shape (Fig. 1A). On MEA, the strain achieved a diameter of 36‒37 mm after 28 days at 25℃. The colonies on MEA appeared olivaceous-black and were velvety with entire margins (Fig. 1B). The conidiophores were ovoid to ellipsoidal, oblong, and apically branched, with either aseptate or 1-septate hyphal cells that were constricted at the septa and brown to pale olivaceous in color (Fig. 1C and 1D). The terminal conidia frequently appeared ovoid and smaller than the other conidia within the chains, suggesting blastic, acropetal conidiogenesis (Fig. 1E). Conidia were 3.2‒5.8 × 2.0‒3.0 µm (n=30) in diameter, pale olivaceous to brown in color, oblong to subglobose and ellipsoidal in shape, smooth, and mostly aseptate, with the conidial chains infrequently branched (Fig. 1F). The cultural and morphological characteristics of the isolated strain KNUF-23-321L indicated that it is most likely related to C. floridana strain SR3028T (Table 2) [6].
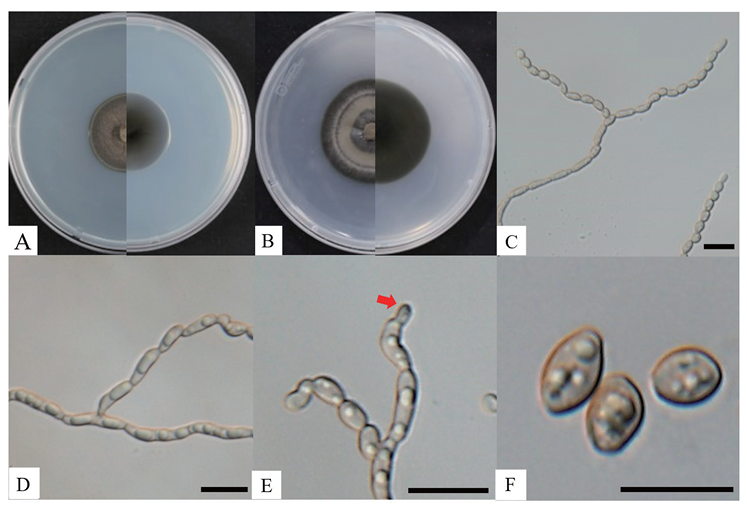
Fig. 1. Cultural and morphological characteristics of Cladophialophora floridana KNUF-23-321L. Cultures were grown at 25℃ for 28 days. A: front and reverse view of the colony on PDA; B: front and reverse view of the colony on MEA; C: aseptate or 1-septate conidiophores; D: chain conidia branched from conidiophore; E: arrow points to terminal conidia exhibiting an ovoid shape, suggesting blastic, acropetal conidiogenesis; F: conidia. Scale bars = 10 µm.
Table 2. Comparison of the cultural and morphological characteristics of strains KNUF-23-321L and Cladophialophora floridana SR3028T
| Characteristic | Cladophialophora floridana KNUF-23-321La | Cladophialophora floridana SR3028Tb |
|---|---|---|
| Colony on PDA | 32‒34 mm, velvety, dry, margin entire, radially folded, olivaceous-black | 22‒23 mm, velvety, dry, margin entire, radially folded, olivaceous-black |
| Colony on MEA | 36‒37 mm, velvety, dry, margin entire, radially folded, olivaceous-black | 22‒23 mm, velvety, dry, margin entire, radially folded, olivaceous-black |
| Conidiophores | Ovoid, ellipsoidal, oblong, and apically branched, aseptate or 1-septate, born terminally on hyphae, brown to pale olivaceous | Oblong to cylindrical, aseptate or 1-septate, born terminally on hyphae, brown to pale olivaceous |
| Conidia | 3.2‒5.8 × 2.0‒3.0 μm, smooth, thick walled, oblong to subglobose, conidial chains sparsely branched, mostly aseptate, pale olivaceous to brown | 3.5‒7.9 × 2.0‒3.2 μm, smooth, thick walled, oblong to subglobose, conidial chains sparsely branched, mostly aseptate, pale olivaceous to brown |
PDA: potato dextrose agar; MEA: malt extract agar.
T Type strain; a Fungal strain used in this paper; b Source of descriptions [6].
The ITS regions and LSU gene sequences were amplified to obtain 573 bp and 815 bp, respectively, for the sequencing analyses of strain KNUF-23-321L. The ITS region showed a high similarity of 98.6% with C. floridana SR3028T and a similarity of 94.3% with C. tortuosa BA4b006. The LSU gene sequence was identical (100% similarity) with the C. floridana SR3028T and showed a similarity of 99.4% with C. tortuosa BA4b006. Based on the above results, the ITS region and LSU gene sequences were combined to create an NJ phylogenetic tree (Fig. 2), in which strain KNUF-23-321L clustered together with C. floridana SR3028T. Based on the morphological characteristics and phylogenetic analysis, strain KNUF-23-321L was identified as C. floridana, an unreported species in Korea.
The genus Cladophialophora was established to unite fungal species that exhibit most of the characteristics of Cladosporium but Phialophora-like conidiogenesis [6]. Although C. floridana KNUF23-321L is closely related to C. tortuosa phylogenetically, it can be differentiated by the conidia and conidiophores. In C. floridana, the apical cells of conidiophore are straight, while those of C. tortuosa are distinctly bent [6]. To date, Cladophialophora has 45 recorded species with diverse habitats. While at least 11 Cladophialophora species are considered opportunistic pathogens that can be found infecting humans and other mammals, the species are also known to be sampled from polluted environmental sources, such as hydrocarbon-rich soil [6,19]. However, some species of the genus Cladophialophora have also been reported to exist as plant endophytes [11,20]. Though there are no reports of pathogenicity from C. floridana, it is thought that other species in the genus Cladophialophora may also be pathogenic, and thus further research on this genus is necessary.
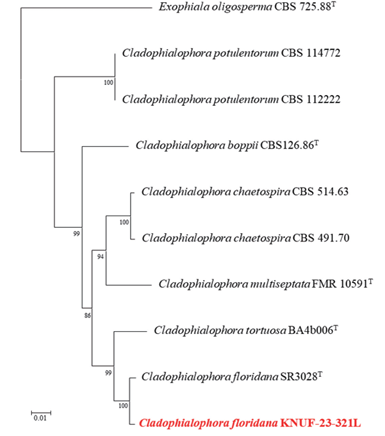
Fig. 2. Neighbor-joining phylogenetic tree based on a combined dataset of partial sequences of internal transcribed spacer (ITS) regions and large subunit (LSU) gene showing the phylogenetic position of strain Cladophialophora floridana KNUF-23-321L among Cladophialophora species and its closest relationship with C. floridana. Bootstrap values greater than 80% (percentage of 1,000 replications) are shown at branching points. The strain isolated in this study is in bold and red. The tree was rooted using Exophiala oligosperma CBS 725.88T as an out-group. Bar, 0.01 substitutions per nucleotide position.
On PDA, the colonies grew to 16‒22 mm in diameter after 20 days at 25℃. The colonies appeared grey to grey-brown on the obverse side and pale brown on the reverse side, with wavy pale margins (Fig. 3A). Setae were observed to be erect, cylindrical, branched, straight, and brown in color, with a length of 59.9‒75.9 × 2.2‒2.8 µm (Fig. 3B). Chlamydospores were dark brown in color and thick-walled (Fig. 3C). Conidia were 2.8‒4.0 × 2.3‒3.4 µm (n = 30) in diameter, hyaline, ellipsoidal, and aseptate (Fig. 3D). The cultural and morphological characteristics of the isolated strain KNUF-23-314 indicated that it is most likely related to Ch. setosum CGMCC 3.20741T (Table 3).
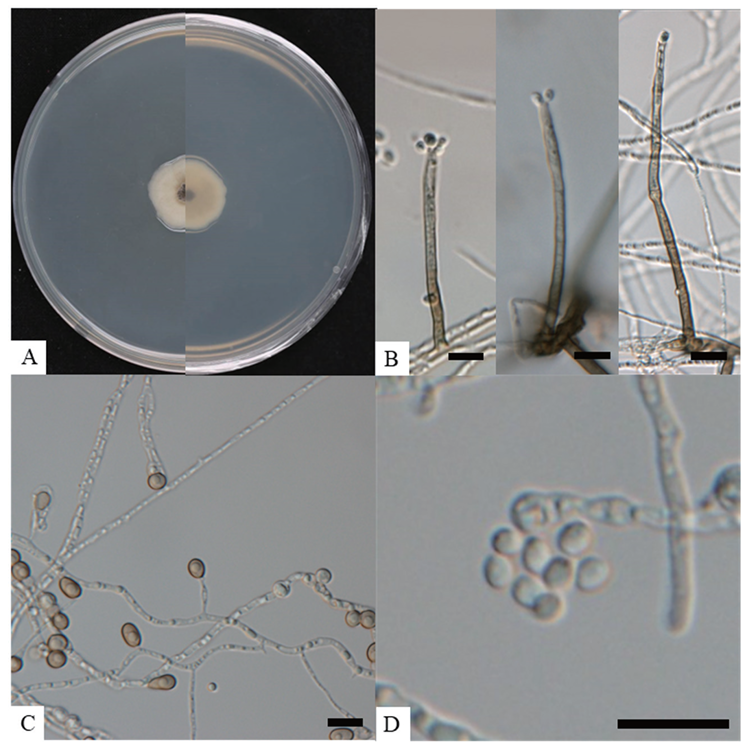
Fig. 3. Cultural and morphological characteristics of Chloridium setosum KNUF-23-314. Cultures were grown at 25℃ for 20 days. A: front and reverse view of the colony on PDA; B: various lengths of setae; C: terminal chlamydospores; D: conidia accumulating in slimy whitish heads. Scale bars = 10 μm.
Table 3. Comparison of the cultural and morphological characteristics of strains KNUF-23-314 and Chloridium setosum CGMCC 3.20741T
| Characteristic | Chloridium setosum KNUF-23-314a | Chloridium setosum CGMCC 3.20741Tc |
|---|---|---|
| Colony on PDA | 16‒22 mm, grey to grey-brown, with pale margins, reverse pale brown | 15‒20 mm, grey to grey-brown, with pale margins, reverse pale brown to dark brown |
| Conidia | 2.8‒4.0 × 2.3‒3.4 μm, hyaline, ellipsoidal, aseptate | 3‒3.8 × 2‒2.5 μm, hyaline, ellipsoidal, subglobose, aseptate |
| Setae | 59.9‒75.9 × 2.2‒2.8 μm, cylindrical, branched, erect, straight, brown | 80‒137 × 2.5‒3.7 μm, cylindrical, branched, erect, straight, brown to dark brown |
PDA: potato dextrose agar
T Type strain; a Fungal strain used in this paper; c Source of descriptions [10].
The ITS regions and LSU gene sequences were amplified to obtain 513 bp and 788 bp, respectively, for the sequencing analyses of strain KNUF-23-314. The ITS region showed a 100% similarity with that of Chloridium setosum CBS 263.76A, a similarity of 98.2%. with Ch. virescens CBS 145487, and a similarity of 96.9% with Ch. bellum var. bellum CBS 709.73A. The LSU gene sequence of the strain was also identical to that of Ch. setosum CBS 263.76A, confirming its position in the genus Chloridium. Based on the above results, the concatenated ITS region and LSU gene sequences were used to generate an NJ phylogenetic tree (Fig. 4). The tree shows strain KNUF-23-314 clustering with Ch. setosum CBS 263.76A. Based on the morphological characteristics and phylogenetic analysis, strain KNUF-23-314 was identified as Ch. setosum, a species previously unreported in Korea.
The genus Chloridium has been isolated from various parts of the world. Most species are found in temperate, subtropical, and tropical regions, with only a few species inhabiting boreal and tundra regions [10]. As such, the genus Chloridium is temperature sensitive, with temperature determining its habitat and geographical distribution. In the case of Ch. setosum, since it was first discovered in tree remnants in China, it has been found worldwide in a variety of environments, including forests, grasslands, woodlands, and agricultural areas, but not in boreal or tundra regions [11]. Fungi of the genus Chloridium form a secondary metabolite, javacin, which is known to have antagonistic effects on soilborne plant pathogens such as Rhizoctonia solani and Verticillium dahliae [16]. Javacin can also be used to control diseases caused by Pseudomonas spp. in humans, animals, and various plants [10] and has been reported to control diseases caused by R. solani, V. dahliae, and Cercospora arachidicola [21].
Given that only two species of the genus Chloridium, namely Ch. virescens and Ch. apiculatum, have been documented in Korea from soil samples, the isolation of Ch. setosum for the first time holds particular importance as it has not been previously reported in Korea. Consequently, this research endeavor has the potential to enrich the fungal biodiversity in Korea by adding a new species to the existing species inventory.
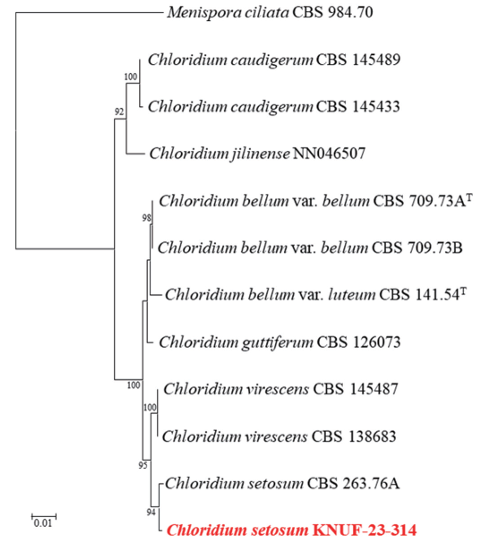
Fig. 4. Neighbor-joining phylogenetic tree based on a combined dataset of partial sequences of internal transcribed spacer (ITS) regions and large subunit (LSU) gene showing the phylogenetic position of strain Chloridium setosum KNUF-23-314 among Chloridium species and its closest relationship with Ch. setosum. Bootstrap values greater than 90% (percentage of 1,000 replications) are shown at branching points. The strain isolated in this study is in bold and red. The tree was rooted using Menispora ciliata CBS 984.70 as an out-group. Bar, 0.01 substitutions per nucleotide position.
The authors declare that there are no conflicts of interest.
This study received financial backing from a grant provided by the National Institute of Biological Resources (NIBR), which is supported by the Ministry of Environment (MOE) of the Republic of Korea (NIBR202304104)
1. Ahn JH, Kim BY, Kim DH, Song JK, Weon HY. Application of amplicon pyrosequencing in soil microbial ecology. Korean J Soil Sci 2012;45:1073-85. [DOI]
2. Frąc M, Hannula SE, Bełka M, Jędryczka M. Fungal biodiversity and their role in soil health. Front Microbiol 2018;9:1-9. [DOI]
3. Bhunjun CS, Niskanen T, Suwannarach N, Wannathes N, Chen YJ, McKenzie EHC, Maharachchikumbura SSN, Buyck B, Zhao CL, Fan YG, et al. The numbers of fungi: are the most speciose genera truly diverse? Fungal Divers 2022;114:387-462. [DOI]
4. Haase G, Sonntag L, Melzer-Krick B, de Hoog GS. Phylogenetic inference by SSU gene analysis of members of the Herpotrichiellaceae, with special reference to human pathogenic species. Stud Mycol 1999;43:80-97.
5. Badali H, Gueidan C, Najafzadeh MJ, Bonifaz A, van den Ende AHGG, de Hoog GS. Biodiversity of the genus Cladophialophora. Stud Mycol 2008;61:175-91. [DOI]
6. Obase K, Douhan GW, Matsuda Y, Smith ME. Cladophialophora floridana and Cladophialophora tortuosa, new species isolated from sclerotia of Cenococcum geophilum in forest soils of Florida, USA. Mycoscience 2015;57:26-34. [DOI]
7. Crous PW, Schubert K, Braun U, de Hoog GS, Hocking AD, Shin HD, Groenewald JZ. Opportunistic, human-pathogenic species in the Herpotrichiellaceae are phenotypically similar to saprobic or phytopathogenic species in the Venturiaceae. Stud Mycol 2007;58:185-217. [DOI]
8. Madrid H, Hernandez-Restrepo M, Gené J, Cano J, Guarro J, Silva V. New and interesting chaetothyrialean fungi from Spain. Mycol Prog 2016;15:1179-201. [DOI]
9. Das K, Lee SY, Jung HY. Cladophialophora lanosa sp. nov., a new species isolated from soil. Mycobiology 2019;47:173-9. [DOI]
10. Réblová M, Hernández-Restrepo M, Sklenář F, Nekvindová J, Réblová K, Kolařík M. Consolidation of Chloridium: new classification into eight sections with 37 species and reinstatement of the genera Gongromeriza and Psilobotrys. Stud Mycol 2022;103:87-212. [DOI]
11. Wu W, Diao Y. Anamorphic chaetosphaeriaceous fungi from China. Fungal Divers 2022;116:1-546. [DOI]
12. Park S, Ten L, Lee SY, Back CG, Lee JJ, Lee HB, Jung HY. New recorded species in three genera of the Sordariomycetes in Korea. Mycobiology 2017;45:64-72. [DOI]
13. Gardes M, Bruns TD. ITS primers with enhanced specificity for basidiomycetes-application to the identification of mycorrhizae and rusts. Mol Ecol 1993;2:113-8. [DOI]
14. White TJ, Bruns TD, Lee SB, Taylor JW. Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics. In: Innis MA, Gelfand DH, Sninsky JJ, editors. PCR protocols: a guide to methods and applications. San Diego: Academic Press; 1990. p. 315-22. [DOI]
15. Vilgalys R, Hester M. Rapid genetic identification and mapping of enzymatically amplified ribosomal DNA from several Cryptococcus species. J Bacteriol 1990;172:4238-46. [DOI]
16. Saitou N, Nei M. The neighbor-joining method: A new method for reconstructing phylogenetic trees. Mol Biol Evol 1987;4:406-25.
17. Kumar S, Stecher G, Li M, Knyaz C, Tamura K. MEGA X: Molecular evolutionary genetics analysis across computing platforms. Mol Biol Evol 2018;35:1547-9. [DOI]
18. Kimura M. A simple method for estimating evolutionary rates of base substitutions through comparative studies of nucleotide sequences. J Mol Evol 1980;16:111-20. [DOI]
19. Blasi B, Tafer H, Kustor C, Poyntner C, Lopandic K, Sterflinger K. Genomic and transcriptomic analysis of the toluene degrading black yeast Cladophialophora immunda. Sci Rep 2017;7:1-13. [DOI]
20. de Hoog G, Nishikaku AS, Fernandez-Zeppenfeldt G, Padín-González C, Burger E, Badali H, Richard-Yegres N, van den Ende AHGG. Molecular analysis and pathogenicity of the Cladophialophora carrionii complex, with the description of a novel species. Stud Mycol 2007;58:219-34. [DOI]
21. Kharwar RN, Verma VC, Kumar A, Gond SK, Harper JK, Hess WM, Lobkovosky E, Ma C, Ren Y, Strobel GA. Javanicin, an antibacterial naphthaquinone from an endophytic fungus of neem, Chloridium sp. Curr Microbiol 2009;58:233-8. [DOI]