Chang Sun Kim*, Young-Nam Kwag, Sang-Kuk Han, and Dae Ho Kim
Forest Biodiversity Division, Korea National Arboretum, Pocheon 11186, Korea
*Correspondence to changsun84@korea.kr
Korean Journal of Mycology (Kor J Mycol) 2024 September, Volume 52, Issue 3, pages 177-187.
https://doi.org/10.4489/kjm.520303
Received on June 17, 2024, Revised on September 03, 2024, Accepted on September 04, 2024, Published on Sep 30, 2024.
Copyright © The Korean Society of Mycology.
This is an Open Access article which is freely available under the Creative Commons Attribution-NonCommercial 4.0 International License (CC BY-NC) (https://creativecommons.org/licenses/by-nc/4.0/).
During a survey of the Korean macrofungal diversity, we identified three Peziza species that have not been previously observed in the Republic of Korea. Phylogenetic and morphological investigations were performed to confirm the taxonomic placement of these species. Herein, three Peziza species (P. granularis, P. saniosa, and P. varia) have been described as newly identified species in the Republic of Korea.
Peziza granularis, Peziza saniosa, Peziza varia, morphology, phylogeny, taxonomy
Macrofungi are broadly classified into Basidiomycetes and Ascomycetes. We were interested in species belonging to the Ascomycetes group, specifically to the genus Peziza. Owing to its small size and inconspicuous appearance, it is one of the macrofungi that most people are unfamiliar with Peziza, the type genus of the family Pezizaceae, was first described by Dillenius in 1719. It is primarily distributed from the temperate to polar regions and is almost absent in tropical regions [1]. In addition, Peziza species thrive in various environments. Specifically, Peziza domiciliana is known for its ability to grow in a wide range of environments [2]. This species flourishes in damp environments, such as plaster, sand, coal dust, refrigerators, water-leakage prone areas, basements and so on.
By 2008, 104 species had been recorded within the Peziza genus worldwide [3]. Among them, five species of Peziza reported in the Republic of Korea (P. domiciliana, P. echinospora, P. praetervisa, P. repanda, and P. vesiculosa; refer to ʻIndex of Korean Mushroom,’ http://www.nature.go.kr/kfni/index.do). However, microscopic observational records, specimens, and molecular phylogenetic analyses of Korean Peziza species are lacking. Therefore, further collection and taxonomic reviews of these species are necessary.
Significant taxonomic changes have occurred worldwide in recent years due to active molecular phylogenetic fungal analyses. Extensive molecular phylogenetic studies of the genus Peziza were published in 2001 [4]. The DNA region utilized belongs to the ribosome (e.g. internal transcribed spacer, large subunit and small subunit of rDNA) the cellular organelle responsible for protein synthesis (e.g. RNA polymerase II and β-tubulin), and reveals seven major groups within the genus Peziza. Subsequent research focused on the core group containing the type species of Peziza, Peziza vesiculosa, utilizing sequences from various protein-coding regions to differentiate species more accurately [5]. Consequently, what was previously considered the genus Peziza has now been divided into nine genera (Peziza, Lepidotia, Sarcopeziza, Inopezia, Malvipezia, Elaiopezia, Paragalactinia, Phylloscypha, and Legaliana) [6].
Most species in the genus Peziza are considered inedible because of their rubbery texture. However, apart from reports of toxicity of the poisonous cup fungus (P. vesiculosa), specific information regarding the toxicity of Peziza species is lacking [2]. Nevertheless, two cases of hypersensitivity pneumonitis related to Peziza species have been reported. In the first case, a previously healthy woman experienced severe respiratory distress and was diagnosed with restrictive lung disease and alveolitis. The fruiting bodies of some fungi were found in the basement, which had been flooded by heavy rain, and air sampling confirmed the presence of Peziza spores [7]. The second case involved a carnation cutter who developed hypersensitivity pneumonitis attributable to Peziza ostracoderma exposure. Although cases of higher concentrations of P. vesiculosa spores in the airways of patients with asthma have been reported, whether this species contributes to asthma onset remains unexplored [8].
This study aimed to clarify the phylogenetic placement of the four Peziza specimens collected during a survey conducted in 2021. In addition, we have described the four Peziza species that have not been previously reported in the Republic of Korea and designated them with Korean names.
The four Peziza specimens used in this study are listed in Table 1. The dried specimens were deposited in the herbarium of the Korea National Arboretum (KH). Macromorphological descriptions were taken from field notes and color photographs of the apothecia. Micromorphological data were obtained from the dried specimens using a light microscope after sectioning and rehydration. Spore size was determined by measuring ca. 30 mature spores.
Table 1. Details of Peziza specimens used in the phylogenetic analyses
| Scientific name | Voucher/Isolate | GenBank No. | Origin |
|---|---|---|---|
| Paragalctinia succosella | KH-97-139 (C) | DQ200841 | Denmark |
| Peziza alcis | s.n. (H, holotype) HOLOTYPE | AF491612 | Finland |
| DHP 11-691 | JQ654491 | USA | |
| KH 00.019 (C) | AF491611 | Norway | |
| Peziza ammophila | AMB 17106 EPITYPE | KX271736 | Italy |
| KH-98-88 (C) | AF491622 | Denmark | |
| L 920528 | KX271724 | Netherlands | |
| Peziza aff. ammophila | KA21-1197 | PP854535 | South Korea |
| L 789131 | KX271731 | Hungary | |
| NSW 7081 (OSC) | AF491621 | USA | |
| Peziza ampliata | TAAM 062922 | JQ654492 | Russia |
| Peziza arvernensis | KS-95-09 (C) | AF491583 | Denmark |
| L 9985 (L) | AF491581 | Netherlands | |
| Peziza azureoides | MPU:JCD 856-75 HOLOTYPE | MT278876 | France |
| Peziza buxoides | MPU:JCD 288-74 HOLOTYPE | MT278880 | France |
| Peziza domiciliana | 16472 | JF908561 | Italy |
| Peziza echinospora | 11663 | JF908533 | Italy |
| Jukka Vauras 9110F (TURA) | AF491575 | Finland | |
| NSW 6763 (OSC) | AF491574 | USA | |
| Peziza fimeti | EB 050691-23 | JQ654488 | Italy |
| Peziza fruticosa | AMB 17135 HOLOTYPE | NR_158836 | Italy |
| Peziza granularis | hr 291 | OP803122 | China |
| 16441 | JF908558 | Italy | |
| KA21-1188 | PP854536 | South Korea | |
| Peziza heimii | MPU:JCD 174-78 | MT635329 | France |
| Peziza hellenica | AMB 17117 HOLOTYPE | KX271723 | Greece |
| Peziza lohjaoensis | s.n. (H, holotype) HOLOTYPE | AF491576 | Finland |
| Peziza luticola | MPU:JCD 489-77 HOLOTYPE | MT278885 | France |
| Peziza martinicensis | LY NV2001.01.01 HOLOTYPE | NR_172791 | France |
| Peziza moseri | 11664 | JF908534 | Italy |
| Peziza nivalis | NSW 6740 (OSC) | AF491619 | USA |
| PDD:72153 | JX845425 | New Zealand | |
| Peziza nivis | MPU:JCD 215-78 HOLOTYPE | MT278887 | France |
| Peziza oceanica | PDD:68977 HOLOTYPE | JX845424 | New Zealand |
| Peziza ostracoderma | DAOM196839 | AY818334 | Unknown |
| F19 | JN002180 | China | |
| Peziza pseudoviolacea | MPU:JCD 52-77 HOLOTYPE | MT278891 | France |
| Peziza saniosa | HBAU15097 | OL441631 | China |
| HMJAU27325 | KU061018 | China | |
| KA21-0301 | PP854537 | South Korea | |
| Peziza subcitrina | KH 00.023 (C) | AF491627 | Denmark |
| KH-97-133 (C) | AF491628 | Denmark | |
| Peziza sublaricina | MPU:JCD 120-78 NEOTYPE | MT278892 | France |
| Peziza varia | KH 00.032 | AF491545 | Denmark |
| KH-97-107 (C) | AF491549 | Denmark | |
| KH-97-88 (C) | AF491556 | Denmark | |
| NSW 6504 (OSC) | AF491565 | USA | |
| TL-4604 (C) | AF491554 | Russia | |
| KA16-1061 | MK351699 | Kyrgyzstan | |
| KA17-0622 | MK351709 | Kyrgyzstan | |
| KA21-0096 | PP854538 | South Korea | |
| Peziza vesiculosa | C no. 52150 (C) | AF491624 | England |
| JV 95.652 (C) | AF491626 | Denmark |
Bold-letters indicated Korean Peziza specimens.
DNA was isolated from fresh fruiting bodies (approximately 0.1 g) using a modified version of the cetrimonium bromide (CTAB) procedure reported by Doyle and Doyle [9]. For species confirmation, the internal transcribed spacer (ITS) region was sequenced using the primer pairs ITS5/ITS4 [10]. PCR mixtures contained 0.5 pmol of each primer, 0.25 mM dNTPs, 10 mM Tris-HCl, 50 mM KCl, 1.5 mM MgCl2 , 2.5 U of Taq DNA polymerase, and 15 ng of template DNA. PCR conditions for ITS was as follows: an initial denaturation step at 94℃ for 4 min, followed by 34 cycles of 94℃ for 40 s, 52℃ for 40 s, and 72℃ for 60 s and a final elongation step at 72℃ for 8 min. The PCR products were purified using an ExoSAP kit (USB, Cleveland, OH, USA). Purified double-stranded PCR fragments were directly sequenced using a BigDye Terminator Cycle Sequencing Kit (Applied Biosystems, Foster City, CA, USA) following the manufacturer’s instructions. Capillary electrophoresis and data collection were performed using the ABI Prism 310 Genetic Analyzer (Applied Biosystems, Foster City, CA, USA). The sequence data were submitted to GenBank (Table 1).
The ITS sequences were aligned using ClustalX 1.81 [11] and trimmed at both terminal ends using Phydit 3.2 [12]. Maximum likelihood (ML) analysis was conducted using PhyML v. 3.0 [13], employing a GTR+G+I model of site substitution with 1,000 bootstrap replicates [14]. Bayesian Inference (BI) analysis was conducted based on the DNA dataset from the results of MrModelTest v. 2.4 [15] using the Markov Chain Monte Carlo (MCMC) algorithm in MrBayes v. 3.1.2 [16]. Two parallel runs were performed using one cold chain and three heated chains for 1 million generations, starting with a random tree. Trees were sampled every 100 generations. We assumed that the two independent runs converged when the average standard deviation (SD) of the split frequencies decreased below 0.05. The trees obtained before convergence were discarded using the burn-in command, and the remaining trees were used to calculate a 50% majority consensus tree and estimate Bayesian posterior probability (BPP). A BPP below 0.95 was not considered significant.
The ITS sequence dataset comprised 509 characters, of which 275 were constant and 193 were parsimony-informative. The estimated base frequencies were as follows: A=0.33358, C=0.19124, G=0.19722, T=0.27797; substitution rates: AC=3.98478, AG=5.17402, AT=4.75843, CG=2.27696, CT=10.25575, GT=1.00000; gamma shape parameter: α=0.909. Paragalactinia succosella was chosen as the outgroup and related genus of Peziza species.
Based on ITS sequence analysis, our specimens represented four species (Peziza ammophila (aff.), P. granularis, P. saniosa, and P. varia). KA21-1188 was in the same group as P. granularis 16441 (Italy) and P. granularis hr 291 (China), but was weakly supported by MLBS and BPP (Fig. 1). However, the ITS sequences of KA21-1188 and two reference strains of P. granularis (P. granularis 16441 and P. granularis hr 291) were almost identical (Fig. 1). Phylogenetically, P. granularis is closely related to P. hellenica AMB 17117 (Greece; Holotype), but they are easily distinguished by morphological characteristics (see taxonomic description part).
Peziza ammophila is a rare species of Asian mycobiota. It grows on sand dunes and beaches, making it easy to distinguish it from other similar brown-colored species. The pseudostipe of this species is fragile and difficult to extract from sand. According to Vizzini et al. [17], their morphological and phylogenetic studies have shown that the P. ammophila complex consists of four subclades (subclades A, B, C and D) [17]. Phylogenetically, our specimen (KA21-1197) belonged to subclade B (refer to P. ammophila L 789131, GenBank No. KX271731; Fig. 1), but we were unable to identify morphological differences from the description of P. ammophila. Vizzini et al. [17] also mentioned this problem and tentatively interpreted them as cryptic species under the provisional name P. deceptive [17]. Therefore, we considered our specimen (KA21-1197) as P. aff. ammophila because of its phylogenetic distinction from the true P. ammophila (subclade A, refer to P. ammophila AMB 17106 Epitype; GenBank No. KX271736) in the ITS tree (Figs. 1 and 2) [17]. Due to the lack of Korean specimens of P. aff. ammophila, more samplings are needed to clarify this status (Figs. 1 and 2).
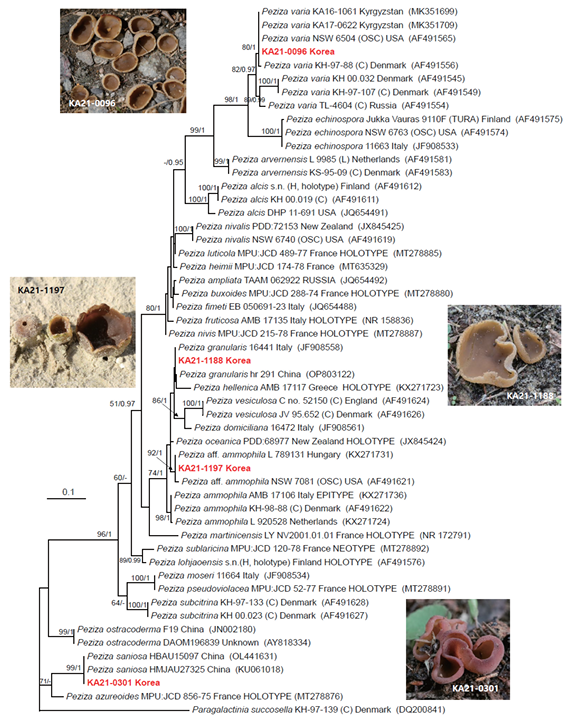
Fig. 1. Phylogram of Peziza species resulting from a maximum likelihood analysis based on ITS sequences. Numbers above or below the branches indicate ML bootstrap support values (MLBS >50%) and Bayesian posterior probabilities (BPP ≥ 0.95). The tree was rooted with Paragalactinia succosella KH-97-139 (C). Specimens from the present study were marked in red. The scale bar indicates the number of nucleotide substitutions per site.

Fig. 2. Peziza aff. ammophila KA21-1197. (A-C) Apothecia; (D-F) Asci and ascospores in 3% KOH; (G) Subhymenium in 3% KOH. Scale bars: A-C=30 mm; D, G=50 μm; E, F=20 μm.
Peziza granularis Donadini ex Van Vooren, Cahiers de la FMBDS 7:56 (2020); Fig. 1 and 3.
Korean name: Ipsang-jubalbeoseot (입상주발버섯), refers to ʻgranularis,’ which means granular
Apothecia 5‒30 mm diam., 5‒10 mm high, cup-shaped when young, expanding later, hymenium smooth, pale brown to yellowish dirty brown, sometimes olive-brown in the center, outer surface granular, powdery, edges thin, and toothed. Asci 270‒300 × 13.4‒15.6 μm, cylindrical, eight spores arranged uniseriately, and dark or pale brown. Ascospores 19‒22 × 9.5‒11.5 μm, ellipsoid, smooth, hyaline, without oil drops. Subhymenium ≤55 μm thick, textura angularis.
Habitat: On moist and wet soils. Growing singly to gregariously.
Examined specimens: Republic of Korea, Chungcheongnam-do, Taean-gun, Nam-myeon Coll. date 13 Oct. 2021 (KA21-1188)
Notes: Peziza granularis is phylogenetically related to P. hellenica (Fig. 1), but they could be easily distinguished by micromorphological characteristics and habitat (P. hellenica: asci 400‒450 × 20‒22 μm, ascospores 24‒28 × 13‒17 μm; sandy dunes in coastal environments) [17]. Morphologically, this species is similar to P. granulosa, but its ascospores are dropless, the ascus width is narrower (P. granulosa: asci 300 × 17‒20 μm), the outer surface is scaleless, and it grows in moist places [18].

Fig. 3. Peziza granularis KA21-1188. (A-C) Apothecia; (D-F) Asi and ascospores in 3% KOH; (G) Subhymenium in 3% KOH. Scale bars: A, B=30 mm; C=10 mm; D=50 μm; E-G=20 μm.
Peziza saniosa J.F. Gmel., Syst. Nat., Edn 13 2(2): 1459 (1792); Figs. 1 and 4.
Korean Name: Geombola-jubalbeoseot (검보라주발버섯), refers to the outer color of apothecia
Apothecia 10‒35 mm diam., 5‒10 mm in high, irregularly cup- to saucer-shaped, expanded, hymenium smooth, dark purple, outer surface pale red-brown, furfuraceous. Asci 300‒410 × 12‒15 μm, cylindrical, eight spores arranged uniseriately, pale brown. Ascospores 14.5‒17.5 × 7.5‒9.5 μm, ellipsoid, scattered coarse warty, hyaline or pale yellow, two oil drops. Subhymenium up to 40 μm thick, textura angularis.
Habitat: Broadleaved and coniferous forests of moss and grass. Grows singly or gregariously.
Examined specimens: The Republic of Korea, Gyeonggi-do, Pocheon-si, Soheul-eup, and Coll. June 28, 2021 (KA21-0301)
Notes: Macroscopically, this species is similar to Geoscypha violacea (Per.) Lambotte (previously Peziza violacea Pers.) and P. depressa Pers. However, they can be distinguished based on their microscopic characteristics. For instance, the ascospores of G. violacea are smooth surface and smaller (13‒13.5 × 7‒7.5 μm) than P. saniosa, and the ascospores of P. depressa are larger (17.5‒20 × 9–11 μm) than P. saniosa, respectively [18].

Fig. 4. Peziza saniosa KA21-0301. (A-C) Apothecia; (D, E) Immature asci in 3% KOH; (F) Asci and ascospores in 3% KOH; (G) Subhymenium in 3% KOH. Scale bars: A-C=20 mm; D=50 μm; E, F=20 μm; G=10 μm.
Peziza varia (Hedw.) Alb. & Schwein., Consp. Fung. (Leipzig): 311 (1805); Fig. 1 and 5.
Korean name: Galsaekkeop-jubalbeoseot (갈색컵주발버섯), refers to its brown-colored apothecia
Apothecia 20‒60 mm diam., 5‒10 mm high, cup- to saucer-shaped, soon irregularly expanded and flat, often stalk existing, margin undulating and weakly notched, hymenium smooth, pale orange, outer surface grey-brownish to pale orange, finely furfuraceous. Asci 250‒300 × 12‒13 μm, cylindrical, eight spores arranged uniseriately, dark brown to pale brown. Ascospores 14.5‒16.8 × 8.1‒10.5 μm, ellipsoid, smooth, sometimes slightly punctate, sometimes one oil drop. Subhymenium up to 50 μm thick, textura angularis.
Habitat: In forests with buried or rotting wood, roots, etc. Grows solely or gregariously.
Examined specimens: Republic of Korea, Gyeonggi-do, Hanam-si, Choi-dong Coll. April 15, 2021 (KA21-0096)
Notes: According to Hansen et al. [1], Peziza varia complex consists of two lineages, based on the ITS sequences of 27 collections. However, they did not find any morphological evidence supporting the division of the two lineages, although there were small differences in spore shape between the two lineages (smooth/finely ornamented). Additionally, no morphological differences of previous descriptions of P. varia were found [1,18]. In our study, our specimen (KA21-0096) was almost identical to the previously reported description of P. varia [1]. Morphologically, P. varia is similar to P. micropus Pers., but the latter species is unlayered or weakly layered and occurs principally on wood (frequently Fagus) [18].
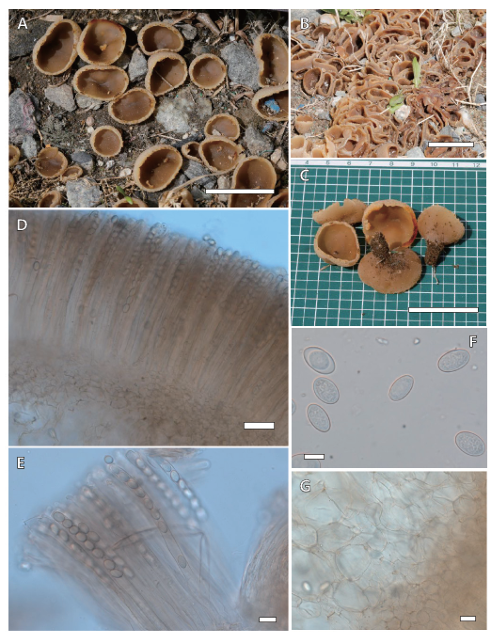
Fig. 5. Peziza varia KA21-0096. (A-C) Apothecia; (D, E) Asci and ascospores in 3% KOH; (F) Ascospores in 3% KOH; (G) Subhymenium in 3% KOH. Scale bars: A-C=30 mm; D=50 μm; E, G=20 μm; F=10 μm.
The authors declare that there are no conflicts of interest.
This study was supported by a research fund from the Korea National Arboretum (project no. KNA1-128-24-1).
1. Hansen K, Læssøe T, Pfister DH. Phylogenetic diversity in the core group of Peziza inferred from ITS sequences and morphology. Mycol Res 2002;106:879-902. [DOI]
2. Arora D. Mushrooms demystified: a comprehensive guide to the fleshy fungi. Berkeley, California: Ten Speed Press; 1986.
3. Kirk PF, Cannon PF, Minter DW, Stalpers JA. Dictionary of the fungi. 10th ed. Wallingford: CABI Publishing; 2008.
4. Hansen K, Læssøe T, Pfister DH. Phylogenetics of the Pezizaceae, with an emphasis on Peziza. Mycologia 2001;93:958-90. [DOI]
5. Hansen K, LoBuglio KF, Pfister DH. Evolutionary relationships of the cup-fungus genus Peziza and Pezizaceae inferred from multiple nuclear genes: RPB2, β-tubulin, and LSU rDNA. Mol Phyl Evol 2005;36:1-23. [DOI]
6. Van Vooren N. Reinstatement of old taxa and publication of new genera for naming some lineages of the Pezizaceae (Ascomycota). Ascomycetes.org 2020;12:179-92.
7. Wright RS, Dyer Z, Liebhaber MI, Kell DL, Harber P. Hypersensitivity pneumonitis from Peziza domniciliana: A case of El Niño lung. Am J Respir Crit Care Med 1999;160:1758-61. [DOI]
8. Madsen AM, Crook B. Occupational exposure to fungi on recyclable paper pots and growing media and associated health effects – A review of the literature. Sci Total Environ 2021;788:147832. [DOI]
9. Doyle JJ, Doyle JL. A rapid DNA isolation procedure for small quantities of fresh leaf tissue. Phytochem Bull 1987;19:11-5.
10. White TJ, Bruns T, Lee S, Taylor J. Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics. PCR protocols: A guide to methods and application. In: Innis DH, Gelfand DH, Sninsky JJ, White TJ, editors. San Diego: Academic Press; 1990. p. 315-22. [DOI]
11. Thompson JD, Gibson TJ, Plewniak F, Jeanmougin F, Higgins DG. The CLUSTAL_X windows interface: Flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res 1997;25: 4876-82. [DOI]
12. Chun J. Computer-assisted classification and identification of actinomycetes [dissertation]. Newcastle upon Tyne: University of Newcastle; 1995.
13. Guindon S, Dufayard JF, Lefort V, Anisimova M, Hordijk W, Gascuel O. New algorithms and methods to estimate maximum-likelihood phylogenies: Assessing the performance of PhyML 3.0. Syst Biol 2010;59:307-21. [DOI]
14. Stamatakis A. RAxML version 8: A tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics 2014;30:1312-3. [DOI]
15. Nouri MT, Lawrence DP, Holland LA, Doll DA, Kallsen CE, Culumber CM, Nylander JAA. MrModeltest v2. Uppsala: Evolutionary Biology Center, Uppsala University; 2004.
16. Ronquist F, Huelsenbeck JP. MrBayes 3: Bayesian phylogenetic inference under mixed models. Bioinformatics 2003;19:1572-4. [DOI]
17. Vizzini A, Lantieri A, Medardi G, Ercole E, Cacialli G. Phylogeny and morphology of the Peziza ammophila complex (Pezizales, Ascomycota), with description of two new species and a new form. Mycol Progress 2016;15:883-901. [DOI]
18. Breitenbach J, Kränzlin F. Fungi of Switzerland Volume 1: Ascomycetes. Lucerne: Verlag Mykologia Luzern; 1984.