Min Kyung Kim, Geon Sik Seo, and Myung Soo Park*
Department of Crops and Forestry, Korea National University of Agriculture and Fisheries, Jeonju 54874, Korea
*Correspondence to mshy1219@af.ac.kr
Korean Journal of Mycology (Kor J Mycol) 2024 September, Volume 52, Issue 3, pages 195-199.
https://doi.org/10.4489/kjm.520305
Received on September 05, 2024, Revised on September 21, 2024, Accepted on September 21, 2024, Published on Sep 30, 2024.
Copyright © The Korean Society of Mycology.
This is an Open Access article which is freely available under the Creative Commons Attribution-NonCommercial 4.0 International License (CC BY-NC) (https://creativecommons.org/licenses/by-nc/4.0/).
Penicillium perform important ecological functions in the rhizosphere, however, only a relatively small number of Penicillium species have been reported from rhizosphere soils. During a study on Penicillium diversity in Korea, we isolated Penicillium strains from the rhizosphere soils of coniferous and broad-leaved trees. Based on phylogenetic analysis of the β-tubulin and calmodulin loci, we accordingly identified two Penicillium species previously unrecorded in Korea: P. nucicola and P. yarmokense. In this paper, we provide comprehensive morphological descriptions of these two species.
Penicillium nucicola; Penicillium yarmokense, Rhizosphere soil, Unrecorded species
The rhizosphere is colonized by a fungal community that plays pivotal roles in plant nutrient absorption and can confer resistance against phytopathogen invasion [1]. Species of the genus Penicillium are established to be among the most common fungi found in rhizosphere soils [2], wherein they influence the stable adaptation of plants to the ecosystem via the production of diverse secondary metabolites, such as solubilized phosphorus, indole acetic acid, siderophores, and extracellular enzymes [3,4]. During a study of Penicillium diversity in Korea, we isolated Penicillium strains from the rhizosphere soil of coniferous and broad-leaved trees. In this study, we identified two Penicillium species from rhizosphere soil previously unrecorded in Korea based on phylogenetic analysis of the ꞵ-tubulin (BenA) and calmodulin (CaM) genes and provided the detailed morphological descriptions.
Rhizosphere soils of coniferous and broad-leaved trees were collected in 2019 and 2020, and fungal strains were isolated on dichloran rose bengal chloramphenicol agar (DRBC; Difco, Becton Dickinson, MD, USA) using the serial dilution method. The isolated Penicillium strains were subsequently cultured on potato dextrose agar (PDA; Difco, Becton Dickinson, MD, USA).
For identification, genomic DNA was isolated from Penicillium strains using an AccuPrep Genomic DNA Extraction Kit (Bioneer, Daejeon, Korea). Each PCR was conducted using a C1000 thermal cycler (Bio-Rad, Richmond, CA), following previously described methods with Bt2a/Bt2b for BenA and CF1/ CF4 or cmd5/cmd6 for CaM [4]. The sequencing was carried out using the PCR primers at Bioneer (Daejeon, Korea). Sequences were manually assembled, proofread, and edited using MEGA5 [5], and have been deposited in the GenBank database (PQ310430-PQ310433 for BenA, PQ310434-PQ310437 for CaM). Sequence similarities for each species were calculated from the two assessed loci using MEGA5 [5]. Multiple sequence alignments were carried out using MAFFT v7 with default settings [6]. Maximum likelihood phylogenetic tree was constructed using RAxML [7] at the CIPRES web portal [8], utilizing the GTR+GAMMA model and 1,000 bootstrap replicates. Isolates AF13142 (NIBRFGC000509171) and AF13692 were accordingly found to cluster as a monophyletic group with the type strain (KAS 2203) of P. nucicola, supported by a 100% bootstrap value. The sequence similarities for BenA ranged from 99.8 to 100%, and was 100% for CaM. Isolates AF12432 (NIBRFGC000509176) and AF12471 were found to group with the type strain (CBS 410.69) of P. yarmokense, supported by a 100% bootstrap value. Sequence similarities for BenA ranged between 99.8 and 100%, and was 100% for CaM (Fig. 1).
The morphological features of the two previously unrecorded species were examined on three different media: Czapek yeast autolysate agar (CYA; Difco, Sparks, MD, USA), malt extract agar (MEA; Oxoid, Hampshire, UK), and yeast extract sucrose agar (YES) [4]. Microscopic characteristics were examined under a light microscope (DM2000; Leica, Germany) using colonies grown on MEA. Color descriptions and alphanumeric codes were based on reference to the Methuen Handbook of Color [9].
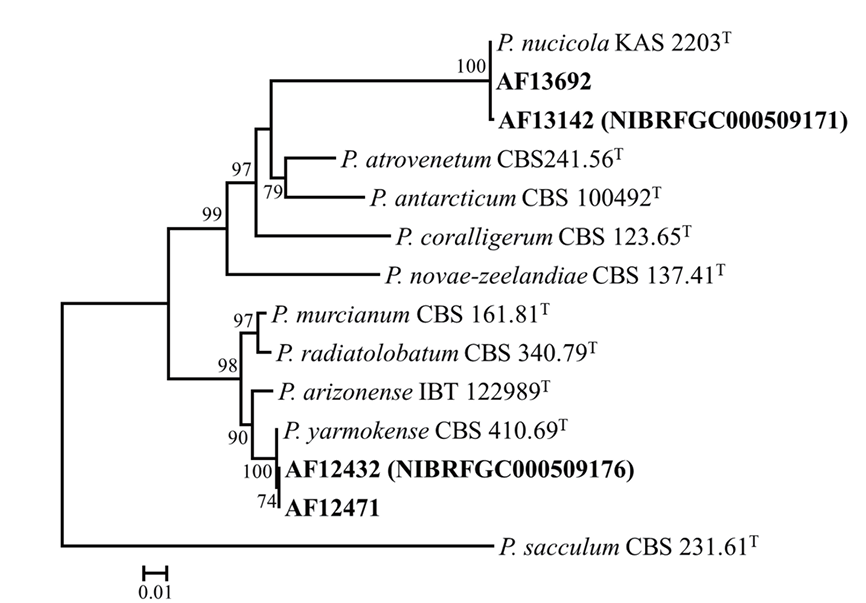
Fig. 1. A maximum likelihood phylogenetic tree based on the concatenated datasets (BenA and CaM) used to identify Penicillium strains isolated from rhizosphere soil. Bootstrap scores of >70 are presented at the nodes. The scale bar indicates the number of nucleotide substitutions per site. “T” indicates the ex-type strains. The strains described in the current study are represented in bold.
Colony diameter after 7 days (in mm): CYA at 25℃: 26-28; CYA at 30℃: no growth; CYA at 37℃: no growth; MEA at 25℃: 30-33; YES at 25℃: 50-51 (Fig. 2).
Colony characteristics: On CYA (25℃ for 7 days): Colonies moderate, radially sulcate; entire, moderately deep, mycelia white at margins; grayish green (28B3) conidia; sporulation moderately dense; texture floccose, with exudates clear; reddish gray soluble pigments; dull green (26E3) to light yellow (4A4) reverse color. On MEA (25℃ for 7 days): Colonies low, plane; entire, low deep at margins; grayish green (27C3) conidia; sporulation moderately dense; texture floccose, with clear exudates; soluble pigments absent; light yellow (3A5) reverse color. On YEA (25℃ for 7 days): Colonies moderate, radially and concentrically sulcate; entire, low deep at margins; sporulation moderately dense; conidia grayish green (26C3); texture floccose, with exudates absent; soluble pigments absent; light yellow (1A4) reverse color.
Conidiophores biverticillate, rough-walled stipes; phialides ampulliform, 7.5‒9.0 × 3.0‒4.0 μm; roughwalled conidia, globose to subglobose, 2.8‒3.3 μm diameter.
Strains examined: Buk-myeon, Ulleung-gun, Gyeongsangbuk-do, Korea, isolated from rhizosphere soil of broad-leaved trees, strains AF13142 (NIBRFGC000509171) and AF13692.
Note: Penicillium nucicola forms a group phylogenetically distinct from other species in the section Canescentia. On all assessed media at 25℃, the Korean strains of P. nucicola were characterized by a more rapid growth compared with that of the type strain [10].
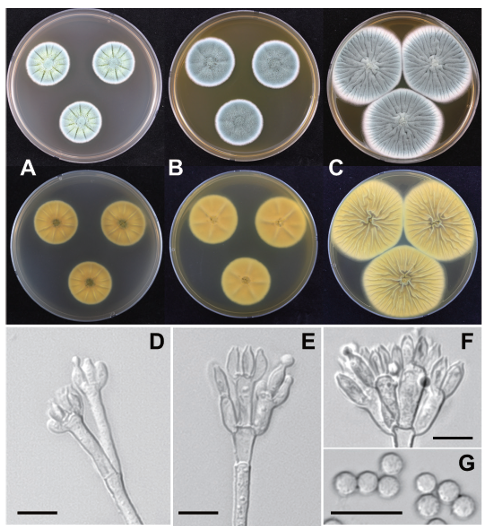
Fig. 2. Penicillium nucicola AF13142 (NIBRFGC000509171) in 7-day-old cultures grown at 25℃. A-C: Colonies grown on Czapek yeast autolysate agar (CYA), malt extract agar (MEA), and yeast extract sucrose agar (YES) from left to right (top=obverse, bottom=reverse). D-F: Conidiophores; G: Conidia (scale bar: D-G=10 μm).
Colony diameter after 7 days (in mm): CYA at 25℃: 27-32; CYA at 30℃: 10-11; CYA at 37℃: no growth; MEA at 25℃: 14-17; YES at 25℃: 23-25 (Fig. 3).
Colony characters: On CYA (25℃ for 7 days): Colonies moderate, radially sulcate; entire, moderately deep, mycelia white at margins; greenish gray (28B2) conidia; sporulation absent to sparse; texture floccose, with clear exudates; soluble pigments absent; pale yellow (3A3) to grayish red (7B4) reverse color. On MEA (25℃ for 7 days): Colonies low, plane; irregular, low at margins; turquoise white (24A2) conidia; sporulation moderately dense; texture floccose, with exudates absent; reddish brown soluble pigments; pale red (8A3) to reddish brown (9A6) reverse color. On YEA (25℃ for 7 days): Colonies moderately deep, radially sulcate; entire, low deep at margins; greenish gray (28B2) conidia; sporulation absent to sparse; texture floccose, with exudates absent; soluble pigments absent; reddish brown (9E7) reverse color.
Conidiophores biverticillate, rough-walled stipes; phialides ampulliform, 7.0‒9.0 × 2.5‒3.0 μm; smooth rough-walled conidia, globose to subglobose, 2.5‒3.0 × 2.4‒3.0 μm diameter.
Strains examined: Naechon-myeon, Hongcheon-gun, Gangwon-do, Korea, isolated from rhizosphere soil of coniferous trees, strains AF12432 (NIBRFGC000509176) and AF12471.
Note: Penicillium yarmokense is phylogenetically similar to P. arizonense in section Canescentia. The former species can be distinguished from the latter by its relatively more rapid growth on MEA and YES [11].
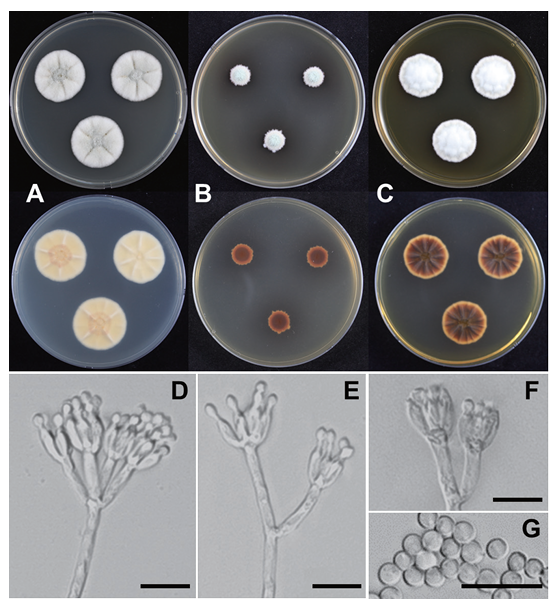
Fig. 3. Penicillium yarmokense AF12432 (NIBRFGC000509176) in 7-day-old cultures grown at 25℃. A-C: Colonies grown on Czapek yeast autolysate agar (CYA), malt extract agar (MEA), and yeast extract sucrose agar (YES) from left to right (top=obverse, bottom=reverse). D-F: Conidiophores; G: Conidia (scale bar: D-G=10 μm).
The authors declare that they have no conflicts of interest.
This research was supported by the Basic Science Research Program through the National Research Foundation of Korea (NRF) funded by the Ministry of Education (No. 2019R1I1A1A01061954) and a grant from the National Institute of Biological Resources (NIBR), funded by the Ministry of Environment (MOE) of the Republic of Korea (NIBR202102107 and NIBR202402104).
1. Pérez-Jaramillo JE, de Hollander M, Ramirez CA, Mendes R, Raaijmakers JM, Carrión VJ. Deciphering rhizosphere microbiome assembly of wild and modern common bean (Phaseolus vulgaris) in native and agricultural soils from Colombia. Microbiome 2019;7: 1-16. [DOI]
2. Workneh F, van Bruggen AHC. Microbial density, composition, and diversity in organically and conventionally managed rhizosphere soil in relation to suppression of corky root of tomatoes. Appl Soil Ecol 1994;1:219-30. [DOI]
3. Altaf MM, Imran M, Abulreesh HH, Khan MS, Ahmad I. Diversity and applications of Penicillium spp. in plant-growth promotion. New and future developments in microbial biotechnology and bioengineering: Penicillum system properties and applications. In: Gupta VK, Rodriguez-Couto S, editors. Amsterdam: Elsevier; 2017. p. 261-76. [DOI]
4. Visagie CM, Hirooka Y, Tanney JB, Whitfield E, Mwange K, Meijer M, Amend AS, Seifert KA, Samson RA. Aspergillus, Penicillium and Talaromyces isolated from house dust samples collected around the world. Stud Mycol 2014;78:63-139. [DOI]
5. Tamura K, Peterson D, Peterson N, Stecher G, Nei M, Kumar S. MEGA5: Molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol Biol Evol 2011;28:2731-9. [DOI]
6. Katoh K, Standley DM. MAFFT multiple sequence alignment software version 7: Improvements in performance and usability. Mol Biol Evol 2013;30:772-80. [DOI]
7. Stamatakis A. RAxML-VI-HPC: Maximum likelihood-based phylogenetic analyses with thousands of taxa and mixed models. Bioinformatics 2006;22:2688-90. [DOI]
8. Miller MA, Pfeiffer W, Schwartz T. Creating the CIPRES science gateway for inference of large phylogenetic trees. SC10 workshop on gateway computing environments (GCE10); 2010 Nov 13-19; New Orleans (LA). Washington DC: IEEE Computer Society; 2010. p. 1-8. [DOI]
9. Kornerup A, Wanscher JH. Methuen handbook of colour. 3rd ed. London: Methuen Publishing; 1978.
10. Visagie CM, Renaud JB, Burgess KM, Malloch DW, Clark D, Ketch L, Urb M, LouisSeize G, Assabgui R, Sumarah MW, et al. Fifteen new species of Penicillium. Persoonia 2016;36:247-80. [DOI]
11. Grijseels S, Nielsen JC, Randelovic M, Nielsen J, Nielsen KF, Workman M, Frisvad JC. Penicillium arizonense, a new, genome sequenced fungal species, reveals a high chemical diversity in secreted metabolites. Sci Rep 2016;6:35112. [DOI]