Yun-Jeong Kim, Jae-Eui Cha, Eun-ju Kim, and Ahn-Heum Eom*
Department of Biology Education, Korea National University of Education, Cheongju 28173, Korea
*Correspondence to eomah@knue.ac.kr
Korean Journal of Mycology (Kor J Mycol) 2024 September, Volume 52, Issue 3, pages 177-187.
https://doi.org/10.4489/kjm.520306
Received on June 17, 2024, Revised on September 03, 2024, Accepted on September 04, 2024, Published on Sep 30, 2024.
Copyright © The Korean Society of Mycology.
This is an Open Access article which is freely available under the Creative Commons Attribution-NonCommercial 4.0 International License (CC BY-NC) (https://creativecommons.org/licenses/by-nc/4.0/).
We isolated endophytic fungi from the leaves of Quercus acutissima Carruth and the stems of Rhododendron mucronulatum Turcz. These fungi were characterized morphologically and subjected to molecular phylogenetic analyses using internal transcribed spacer, large subunit rDNA, translation elongation factor-1-α, the second largest subunit of RNA polymerase II, and β-tubulin sequences. We accordingly identified two endophytic fungi, Didymella coffeae-arabicae and Austropleospora keteleeriae, in the order Pleosporales, which have not previously been recorded in Korea. Herein, we described the morphological characteristics and molecular phylogenetic analyses of these two endophytic fungi, contributing to our understanding of fungal diversity in Korea. Our findings highlight the significance of endophytic fungi in ecological study and their potential utility as a source of novel bioactive compounds.
Austropleospora keteleeriae, Didymella coffeae-arabicae, Endophytic fungus, Pleosporales
The fungal order Pleosporales is the largest within the class Dothideomycetes in the phylum Ascomycota, comprising approximately one-quarter of all known Dothideomycetes species [1]. This order is recognized for its notable diversity, with different species occupying a wide range of ecological niches, including as epiphytes on living leaves and stems, endophytes within plant tissues, and parasites [2, 3]. Additionally, some species function as hyperparasites of other fungi or insects, engage in symbiotic relationships with lichens, and thrive as saprobes on decaying plant material, including stems, leaves, and bark. Most Pleosporales species produce unicellular ascomata [4], and with the exception of those species in the families Diademaceae and Sporormiaceae, these ascomata are characterized by papillate and ostiolate structures [5]. Pseuparaphyses are present within the ascomata [6], and the asci are primarily bitunicate with a double-walled structure. Although generally cylindrical, clavate, or cylindro-clavate, a few species produce inversely obclavate or globose asci [5].
Endophytic fungi are those that colonize plant tissues without causing harm and can potentially confer beneficial properties to the host plant [7], including promoting plant growth, enhancing resistance to diseases, and increasing tolerance to environmental stresses [8]. Additionally, endophytic fungi are a prolific source of secondary metabolites [9], many of which may have potential applications in medicine, agriculture, and industry [10]. Consequently, studies on these fungi can enhance our understanding of the ecological interactions between plants and fungi and provide a rich reservoir of natural compounds for biotechnological exploration.
In this study, we aimed to isolate and characterize previously unreported endophytic fungi in Korea and identified two fungal strains within the order Pleosporales, for which we performed morphological characterization and molecular phylogenetic analyses.
In March 2023, stems of Rhododendron mucronulatum were collected from Samcheok, Gangwon-do, and in April of the same year, leaves of Quercus acutissima were collected from Gongju, Chungcheongnamdo. After confirming the absence of any diseases, the samples were transported to the laboratory within 24 h, where the leaves and stems were washed several times with distilled water and then sequentially surfacesterilized in 35% H2 O2 for 1 min and 70% ethanol for 30 s. The sterilized samples were then cut into segments measuring 1 cm × 0.5 cm (leaves) and 1 cm (stems) and plated on potato dextrose agar (PDA; Difco Laboratories, Detroit, USA). The plates were incubated at 25℃, and fungi growing from within the samples were subcultured to obtain pure cultures.
The isolated fungi were cultured on PDA and malt extract agar (MEA; Kisan bio, Seoul, Korea) at 25℃ for 7 days in the dark for subsequent examination of colony characteristics. The microscopic structures of the cultured fungi were observed using an Axio Imager A2 optical microscope (Carl Zeiss, Overkochen, Germany). DNA was extracted from the fungal mycelia using a HiGene Genomic DNA Prep Kit (BioFACT, Daejeon, Korea) according to the protocol provided by the manufacturer and was used as a template for polymerase chain reaction (PCR) amplification of five different sequence regions for molecular identification. The internal transcribed spacer (ITS) was amplified using ITS1F/ITS4 primers [11], large subunit ribosomal DNA (LSU) using LR0R/LR16 primers [12], β-tubulin (TUB) using Bt2a/Bt2b primers [13], the second largest subunit of RNA polymerase II (RPB2) using fRPB2-5f/fRPB2-7cR primers [14], and translation elongation factor-1-α (TEF) using EF1-983F/EF1-2218R primers [15]. The PCR products obtained were electrophoresed on 1.5% agarose gels for 20 min to confirm the size of the amplified DNA, followed by Sanger sequencing (SolGent, Daejeon, Korea). The DNA sequences were analyzed for similarity with database accessions using the Basic Local Alignment Search Tool of the National Center for Biotechnology Information (NCBI). Phylogenetic trees were constructed using the neighbor-joining method with the MEGA11 program and were validated by 1000 bootstrap replicates. Information on the newly recorded fungal species has been deposited at the National Institute of Biological Resources (NIBR), and the DNA sequences have been deposited in the NCBI database.
Morphological characteristics: Colonies were grown on PDA and MEA media at 25 °C for 7 days, and their characteristics were observed. On PDA, the colonies were circular, flat, and measured 60–66 mm in diameter with a filamentous margin. The center was olive-green in color, gradually fading to an ivorycolored margin. The reverse side of the colony was dark brown to black in the center and ivory-colored at the margin (Fig. 1, Table 1). On MEA, the colonies were circular, flat, and measured 65–74 mm in diameter with a filamentous margin. The center was a beige to yellowish ochre color, and the margin was ivorycolored. The color of the reverse side was similar to that of the front side. Conidia were oval to elliptical in shape, hyaline, and measured 5.45 (4.12–7.4) × 3.67 (2.49‒4.75) μm (n=20). Chlamydospores had thick brown cell walls, are multicellulares and intercalares. Conidiomata exude black to light apricot or beige exudates, typically solitary, with variable sizes and shapes. They are mostly solitary, glabrous, papillate, or possess an elongated neck.
Specimen examined: Gongju, Chungcheongnam-do, Korea, 36°20ʹ9.7ʹʹN, 127°11ʹ14.64ʹʹE, April 28, 2023, Didymella coffeae-arabicae, isolated from the leaves of Quercus acutissima Carruth, strain KNUE23N733, NIBRFGC000520715, GenBank No. PQ199317 (ITS), PQ199319 (LSU), PQ227105 (TUB), and PQ227103 (RPB2)
Notes: Didymella coffeae-arabicae was first reported as Phoma coffeae-arabicae in 2009 [16], but Phoma is phylogenetically polyphyletic, requiring reclassification [17]. Based on phylogenetic analysis of Phoma species using a combination of multiple DNA gene regions, the species was later placed within the genus Didymella in 2015 [18]. Didymella coffeae-arabicae has been reported as an endophyte of various plant species and has been shown to inhibit the pathogen Fusarium oxysporum in the medicinal plant Hydrocotyle verticillata Thunb. [19]. The ITS sequence of the new isolate showed 100% identity with D. coffeae-arabicae CBS 123380 (MH863293), and TUB sequence showed 100% identity with D. coffeaearabicae CBS 123380 (FJ427104). The LSU sequence showed 100% identity with D. cof feae-arabicae C461A (MK348029), and RPB2 sequence showed 100% identity with D. cof feae-arabicae CBS 123380 (KT389603). A phylogenetic tree constructed using the neighbor-joining method based on a combined analysis of ITS, LSU, TUB, and RPB2 sequences revealed that the new isolate formed a monophyletic group with D. coffeae-arabicae CBS 123380 (Fig. 2).
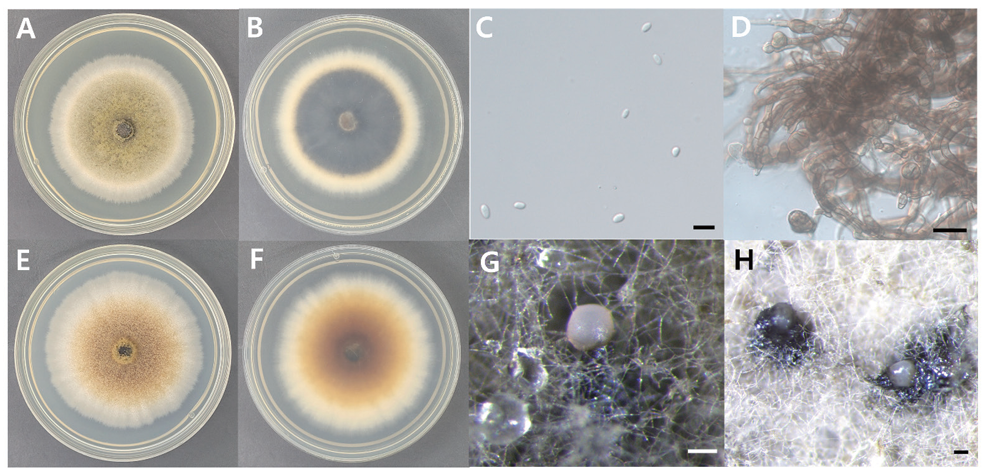
Fig. 1. Morphology of D. coffeae-arabicae KNUE 23N733. A: Colony grown for 7 days on potato dextrose agar (PDA), front. B: Colony grown for 7 days on PDA, reverse. C: Conidia. D: Chlamydospores. E: Colony grown for 7 days on malt extract agar (MEA), front. F: Colony grown for 7 days on MEA, reverse. G, H: Conidiomata. Scale bars: C=10 μm, D=20 μm, G and H=100 μm.
Table 1. Morphological characteristics of the KNUE 23N733 and CBS 123380 strains of Didymella coffeae-arabicae
| Characteristics | D. coffeae-arabicae KNUE 23N733 | D. coffeae-arabicae CBS 123380 [16] |
|---|---|---|
| Colony | PDA, MEA, 25℃, 7 days. | MEA, OA, CHA, 25℃, 7 days. |
| Color | PDA: Front, greenish olivaceous near the center, with ivory edges; reverse, fuscous-black near center, ivory edges. MEA: Front, beige to yellow ochre near the center, with ivory edges; reverse concolorous. | MEA: Front, aerial mycelium white with rosy-vinaceous tinges; agar surface iron-gray; reverse, fulvous to amber, leaden-black in zones with abundant pycnidia. OA: Aerial mycelium white; immersed mycelium hyaline or greenish olivaceous, fuscous-black near center; reverse concolorous. |
| Size | PDA: 60‒66 mm in diameter. MEA: 65‒74 mm in diameter. | PDA: No observations. MEA: 57‒70 mm in diameter. OA: 61‒66 mm in diameter. CHA: Similar growth rate to that on MEA |
| Shape | PDA: Circular, flat, with a filamentous margin. MEA: Circular, flat, with a filamentous margin. | MEA: Entire, smooth, sharp margin with aerial mycelium condensed. OA: Entire, smooth margins with sparse or absent tufted aerial mycelium. CHA: Aerial mycelium compact or tufted, primrose to citrinegreen, pale greenish glaucous near center, leaden-gray near margin; reverse leaden-black. |
| Conidia | Ellipsoidal to ovoid, hyaline, 5.45 (4.12‒7.4) × 3.67 (2.49‒4.75) μm. | Ellipsoidal to ovoid, hyaline, eguttulate, or with 1-4 small polar droplets, 4.5‒6 (4‒7) × 3‒4 (2.5‒4.5) μm. |
| Conidiomata | Black to light apricot to beige exudate, mostly solitary, variable in shape and size, glabrous, papillate or with an elongated neck. | Mostly solitary or in chains, variable in shape and size; mostly ovoid, but also (sub)globose or elongated, glabrous, papillate, or with an elongated neck; mostly uni- or bi-ostiolate, 150‒310 (100‒310) × 110‒200 (100‒240) μm. |
| Chlamydospores | Thick-cell walled, brown, multicellulares, intercalares. | Multicellular, immersed, dictyosporous, pseudosclerotioid, intercalary, or solitary, 40‒100 (23‒190) × 15‒30 (11‒30) μm. |
PDA: potato dextrose agar; MEA: malt extract agar; OA: oatmeal agar; CHA: cherry decoction agar.
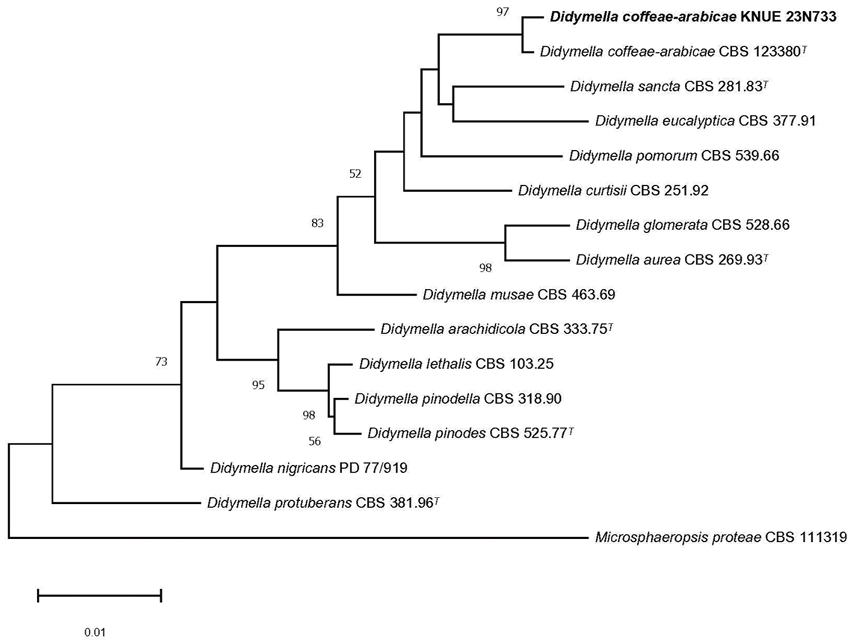
Fig. 2. Neighbor-joining phylogenetic tree of D. coffeae-arabicae KNUE 23N733 based on concatenated sequences of the internal transcribed spacer (ITS), large subunit ribosomal DNA (LSU) regions, β-tubulin (TUB), and the second largest subunit of RNA polymerase II (RPB2). Microsphaeropsis proteae serves as the outgroup. The numbers at the nodes represent bootstrap values greater than 50% (1,000 replicates). Extype strains are indicated by a superscript “T” following the strain reference number. The strain isolated in this study is in a bold.
Morphological characteristics: Colonies were cultivated on PDA and MEA media at 25 °C for 7 days, and their characteristics were observed. On PDA, the colonies were circular, flat, and measured 35–41 mm in diameter with a soft texture. The center of the colonies was beige to light yellow in color, gradually fading toward an ivory-colored margin. The reverse side of the colonies was golden yellow to yellow at the center and light yellow to beige at the margin (Fig. 3, Table 2). On MEA, the colonies were circular, flat, and measured 43–46 mm in diameter with a soft texture. The center was ochre to light yellow, gradually fading toward a white to ivory-colored margin. The reverse side was golden yellow to deep yellow at the center and light yellow to beige at the margin. The conidia are globose to ellipsoid, hyaline or dark brown, and measure 6.26 (5.25‒7.38) × 3.47 (2.37‒4.67) μm (n=20). Conidiomata are dark brown to black, ellipsoidal or elongated ovoid, and glabrous.
Specimen examined: Samcheok-si, Gangwon-do, Korea, 37°12ʹ3.2ʹʹN 129°2ʹ11.2ʹʹE, March 31, 2023, Austropleospora keteleeriae, isolated from the stems of Rhododendron mucronulatum Turcz, strain KNUE23N307, NIBRFGC000518068, GenBank No. PQ199398 (ITS), PQ199399 (LSU), and PQ227104 (TEF).
Notes: The genus Austropleospora was first reported in 2010 with the single species A. osteospermi [20]. In 2015, Paraconiothyrium archidendri was reclassified as a species of Austropleospora [21]. Additional species, including A. keteleeriae in 2012 and A. ochracea in 2019, were subsequently reported, bringing the total to four known species within this genus [22, 23]. A. archidendri has spore-forming cells similar in shape and size to those of A. keteleeriae, but with hyaline, thin-walled spore-forming cells attached to the spore-forming cells. Comparative analysis of the ITS sequences of these two species revealed nine (1.9%) nucleotide differences, identifying A. keteleeriae as a distinct species [24]. The ITS sequence of the new isolated strain showed 99.7% identity with A. keteleeriae ZHKU 22-0209 (OP297802), LSU sequence showed 100% identity with A. keteleeriae ZHKU 22-0209 (OP297772), and TEF sequence showed 99.8% identity with A. keteleeriae MFLUCC 18-1551 (MK360045). A phylogenetic tree constructed using the neighbor-joining method, based on the combined analyses of ITS, LSU, and TEF sequences, revealed that the new isolate formed a monophyletic group with A. keteleeriae MFLUCC 18-1551 (Fig. 4).
In this study, we successfully identified two endophytic fungal species, D. coffeae-arabicae and A. keteleeriae, previously unrecorded in Korea. Both fungi, members of the order Pleosporales, were isolated from different host plants, Q. acutissima and R. mucronulatum, respectively. Morphological and molecular analyses revealed clear distinctions between these fungi and their respective closely related species, confirming their presence in Korea for the first time. The findings of this research substantially contribute to our understanding of fungal biodiversity in Korea and highlight the importance of continued exploration and documentation of endophytic fungi.
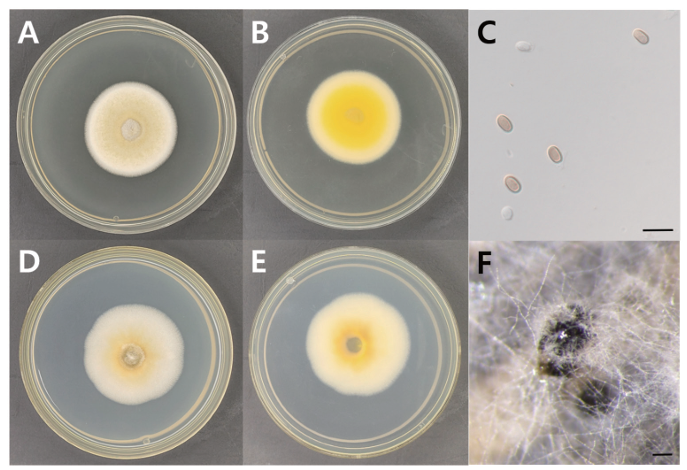
Fig. 3. Morphology of A. keteleeriae KNUE 23N307. A: Colony grown for 7 days on potato dextrose agar (PDA), front. B: Colony grown for 7 days on PDA, reverse. C: Conidia. D: Colony grown for 7 days on malt extract agar (MEA), front. E: Colony grown for 7 days on MEA, reverse. F: Conidiomata. Scale bars: C=10 μm, F=100 μm.
Table 2. Morphological characteristics of the KNUE 23N307 and MFLUCC 18–1551 strains of Austropleospora keteleeriae
| Characteristics | A. keteleeriae KNUE 23N307 | A. keteleeriae MFLUCC 18–1551[23] |
|---|---|---|
| Colony | PDA and MEA, 25℃, 7 days. | MEA, 18℃, 7 days. |
| Color | PDA: Front, pale gold-yellow near the center, with ivory-colored edges; reverse, gold to yellow near the center, ivory-colored edges. MEA: Light yellow to yellow ochre near the center, with ivory to white edges; reverse, gold to dark yellow near the center, ivory to light yellow edges. | MEA: Front, colonies grow in four layers, from the center to margins, showing gray, brown, pinkish gray, and dark brown, respectively; reverse, center brown, middle off-white, and dark brown at margin. |
| Size | PDA: 35‒41 mm in diameter. MEA: 43‒46 mm in diameter. | MEA: 25‒30 mm in diameter. |
| Shape | PDA: Circular, flat, with a filamentous margin, downy. MEA: Circular, flat, with a filamentous margin, downy. | MEA: Circular, with an irregular margin. |
| Conidia | Globose to ellipsoid, hyaline or dark brown, 6.26 (5.25‒7.38) × 3.47 (2.37‒4.67) μm. | Solitary, hyaline when young, becoming dark brown at maturity, globose to obovate, single-celled, thick, and smooth-walled, 5 (4‒5.5) × 5.5 (5‒6) μm. |
| Conidiophores | No observations. | Conidiophores reduced to conidiogenous cells, arising from the base and sides of the conidioma; phialidic, enteroblastic, determinate, ampulliform, lining the inner wall layer of the pycnidium; hyaline, smooth, thin-walled, 6.2 (5‒7) × 4.5 (4‒5) μm. |
| Conidiomata | Dark brown to black, ellipsoidal or elongated ovoid, glabrous. | 210‒240 μm high × 220‒255 μm in diameter ( =228 × 242 μm, n=30); pycnidial, solitary, immersed, globose to obpyriform, unilocular, centrally ostiolate. Wall 33‒55 μm wide ( =44 μm; n=20), thick, 5 or 6 layered, composed of an outer 4 or 5 layers, brown, and inner 1 or 2 layers of hyaline, thin-walled cells of textura angularis |
PDA: potato dextrose agar; MEA: malt extract agar.
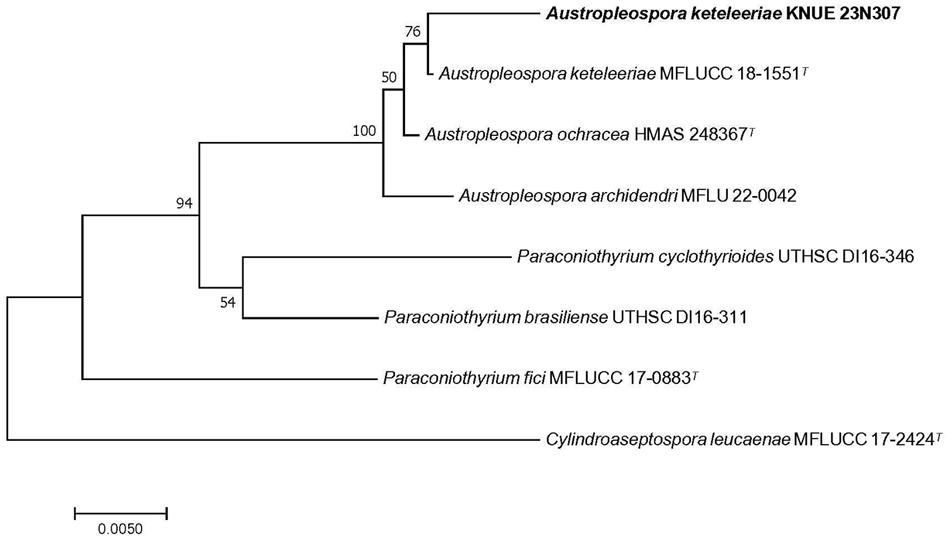
Fig. 4. Neighbor-joining phylogenetic tree of A. keteleeriae KNUE 23N307 based on concatenated sequences of the internal transcribed spacer (ITS), large subunit ribosomal DNA (LSU) regions, and translation elongation factor-1-α (TEF). Cylindroaseptospora leucaenae serves as the outgroup. The numbers at the nodes represent bootstrap values greater than 50% (1,000 replicates). Ex-type strains are indicated by a superscript “T” following the strain reference number. The strain isolated in this study is in a bold.
The authors declare that there are no conflicts of interest.
This work was supported by a grant from the National Institute of Biological Resources (NIBR202402104), funded by the ministry of Environment (MOE) of the Republic of Korea.
1. Kirk P, Cannon P, Minter D, Stalpers J. Dictionary of the fungi. Wallingford: CABI; 2008. [DOI]
2. Kruys Å, Eriksson OE, Wedin M. Phylogenetic relationships of coprophilous Pleosporales (Dothideomycetes, Ascomycota), and the classification of some bitunicate taxa of unknown position. Mycol Res 2006;110:527-36. [DOI]
3. Ramesh C. Loculoascomycetes from India. In: Bhat DJ, editor. Frontiers of Fungal Diversity in India. New Delhi: Apex Printers; 2003. p. 457.
4. Clements F, Shear C. Genera of Fungi. New York: H. W. Wilson Company; 1931.
5. Zhang Y, Crous PW, Schoch CL, Hyde KD. Pleosporales. Fungal Divers 2012;53:1-221. [DOI]
6. Liew EC, Aptroot A, Hyde KD. Phylogenetic significance of the pseudoparaphyses in Loculoascomycete taxonomy. Mol Phylogenet Evol 2000;16:392-402. [DOI]
7. Petrini O. Fungal endophytes of tree leaves. In: Andrews JH, Hirano SS, editors. Microbial Ecology of Leaves. New York: Springer; 1991. p. 179-97. [DOI]
8. Sturz A, Nowak J. Endophytic communities of rhizobacteria and the strategies required to create yield enhancing associations with crops. Appl Soil Ecol 2000;15:183-90. [DOI]
9. Strobel G, Daisy B. Bioprospecting for microbial endophytes and their natural products. Microbiol Mol Biol Rev 2003;67:491-502. [DOI]
10. Zhang HW, Song YC, Tan RX. Biology and chemistry of endophytes. Nat Prod Rep 2006;23:753-71. [DOI]
11. Gardes M, Bruns TD. ITS primers with enhanced specificity for basidiomycetes‐application to the identification of mycorrhizae and rusts. Mol Ecol 1993;2:113-8. [DOI]
12. Moncalvo JM, Lutzoni FM, Rehner SA, Johnson J, Vilgalys R. Phylogenetic relationships of agaric fungi based on nuclear large subunit ribosomal DNA sequences. Syst Biol 2000;49:278-305. [DOI]
13. Glass NL, Donaldson GC. Development of primer sets designed for use with the PCR to amplify conserved genes from filamentous ascomycetes. Appl Environm Microbiol 1995;61:1323-30. [DOI]
14. Liu YJ, Whelen S, Hall BD. Phylogenetic relationships among ascomycetes: evidence from an RNA polymerse II subunit. Mol Biol Evol 1999;16:1799-808. [DOI]
15. Rehner SA, Buckley E. A Beauveria phylogeny inferred from nuclear ITS and EF1-α sequences: evidence for cryptic diversification and links to Cordyceps teleomorphs. Mycologia 2005;97:84-98. [DOI]
16. Aveskamp MM, Verkley GJ, de Gruyter J, Murace MA, Perello A, Woudenberg JH, Groenewald JZ, Crous PW. DNA phylogeny reveals polyphyly of Phoma section Peyronellaea and multiple taxonomic novelties. Mycologia 2009;101:363-82. [DOI]
17. Aveskamp MM, de Gruyter J, Woudenberg JH, Verkley GJ, Crous PW. Highlights of the Didymellaceae: a polyphasic approach to characterise Phoma and related pleosporalean genera. Stud Mycol 2010;65:1-60. [DOI]
18. Chen Q, Jiang J, Zhang G, Cai L, Crous PW. Resolving the Phoma enigma. Stud Mycol 2015;82:137-217. [DOI]
19. Sukarno N, Ginting RCB, Widyastuti U, Darusman LK, Kanaya S, Batubara I, Aryantha INP, Waite M. Endophytic fungi from four Indonesian medicinal plants andtheir inhibitory effect on plant pathogenic Fusarium oxysporum. Hayati J Biosci 2021;28:152-71. [DOI]
20. Morin L, Shivas R, Piper M, Tan Y. Austropleospora osteospermi gen. et sp. nov. and its host specificity and distribution on Chrysanthemoides monilifera ssp. rotundata in Australia. Fungal Divers 2010;40:65-74. [DOI]
21. Ariyawansa HA, Hyde KD., Jayasiri SC, Buyck B, Chethana KT, Dai DQ, Dai YC, Daranagama DA, Jayawardena RS, Lücking R. Fungal diversity notes 111-252-taxonomic and phylogenetic contributions to fungal taxa. Fungal Divers 2015;75:27-274. [DOI]
22. Dissanayake LS, Wijayawardene NN, Samarakoon MC, Hyde KD, Kang JC. The taxonomy and phylogeny of Austropleospora ochracea sp. nov.(Didymosphaeriaceae) from Guizhou, China. Phytotaxa 2021;491:217-29. [DOI]
23. Jayasiri S, Hyde K, Jones E, McKenzie E, Jeewon R, Phillips A, Bhat D, Wanasinghe D, Liu J, Lu Y, Kang JC, Xu J, Karunarathna SC. Diversity, morphology and molecular phylogeny of Dothideomycetes on decaying wild seed pods and fruits. Mycosphere 2019;10: 1-186. [DOI]
24. Jeewon R. Establishing species boundaries and new taxa among fungi: recommendations to resolve taxonomic ambiguities. Mycosphere 2016;7:1669-77. [DOI]