Mohammad Hamizan Azmi1, Seong-Keun Lim1, Sang-Min Lee1, Dae-Hoon Kang1, Jin-Sil Choi1, Gwang-Jae Lim1, Jun-Woo Choi1, Min-Kyu Kim1, Seung-Yeol Lee1,2,*, and Hee-Young Jung1,2
1Department of Plant Medicine, Kyungpook National University, Daegu 41566, Korea
2Institute of Plant Medicine, Kyungpook National University, Daegu 41566, Korea
*Correspondence to leesy1123@knu.ac.kr
Korean Journal of Mycology (Kor J Mycol) 2024 September, Volume 52, Issue 3, pages 211-221.
https://doi.org/10.4489/kjm.520307
Received on September 09, 2024, Revised on September 25, 2024, Accepted on September 25, 2024, Published on Sep 30, 2024.
Copyright © The Korean Society of Mycology.
This is an Open Access article which is freely available under the Creative Commons Attribution-NonCommercial 4.0 International License (CC BY-NC) (https://creativecommons.org/licenses/by-nc/4.0/).
This study reports the isolation and characterization of two previously unrecorded fungal strains, designated KNUF-23-232 and KNUF-23-344. The initial classification was based on an examination of their morphological and cultural characteristics. To further confirm their identities and elucidate their evolutionary relationships, molecular phylogenetic analyses were conducted using sequences from the internal transcribed spacer (ITS) region, the large subunit of 28S rRNA (LSU), the second largest subunit of RNA polymerase II (RPB2), and translation elongation factor-1α (TEF1). The largest subunit of RNA polymerase II (RPB1) was also amplified for strain KNUF-23-344. The cultural and morphological traits of strain KNUF23-232 were consistent with those of Blackwellomyces calendulinus BCC 68502T, whereas those of strain KNUF-23-344 corresponded to Neoaraneomyces araneicola DY101711T. The integration of morphological and phylogenetic analyses led to the identification of KNUF-23232 as B. calendulinus and KNUF-23-344 as N. araneicola. To the best of our knowledge, this study represents the first documentation of B. calendulinus and N. araneicola in Korea.
Blackwellomyces calendulinus, Entomogenous fungi, Morphology, Neoaraneomyces araneicola, Phylogeny
Entomogenous fungi are typically associated with insects, other arthropods, and even non-arthropod microinvertebrates, primarily functioning as pathogens or parasites [1]. Although these fungi and their related species exhibit a wide range of interactions with their hosts, they follow similar biological principles in the diseases they cause. Infection typically occurs through the penetration of the host’s cuticle following a series of competitive interactions, with successful infection requiring the fungi to overcome or evade the host’s defenses [2]. Recent advancements in the phylogenetic classification of fungi have enhanced the understanding of various entomopathogenic fungal taxa, potentially facilitating their unconventional applications and encouraging the use of novel fungal entomopathogens as biocontrol agents. Among these, the genus Blackwellomyces, belonging to the order Hypocreales and family Cordycipitaceae, was first isolated from the larvae of coleopteran and lepidopteran insects, with eight species have been reported worldwide [3,4]. The genus Blackwellomyces was initially established to accommodate B. cardinalis and has Acremonium-like, Evlachovaea-like, and Mariannaea-like conidial arrangements, with multiple forms observed within a single species [4]. Additionally, the genus Neoaraneomyces, belonging to the order Hypocreales and family Clavicipitaceae, was isolated from a dead spider carcass in China and exhibits a conidial chain resembling that of Paecilomyces [5]. Although spiders are not classified as arthropods, a study conducted in 2023 classified the spider-pathogenic fungus Neoaraneomyces as entomogenous [6]. The etymology of the genus reflects its characterization as a new genus of fungi parasitic to spiders, with the type species being Neoaraneomyces araneicola [5]. This study aimed to isolate fungi from domestic soil samples and identify the isolated fungi based on morphological and molecular biological characteristics, with the objective of securing domestic fungal resources and reporting potential endemic species.
Soil samples were collected from Chungnam (36°11’44.1″N, 127°16’24.0″E) and Jeonbuk Province (35° 58’56.3″N, 126°41’46.1″E) in Korea, and used as the source of fungal isolates in this study. The isolation of fungi from the soil samples using the serial dilution technique, as previously described [7]. Colonies showing signs of germination were subsequently transferred to fresh potato dextrose agar (PDA; Difco, Detroit, MI, USA) plates and incubated at 25℃. The fungal strains KNUF-23-232 and KNUF-23-344 were selected for further molecular analyses, as well as cultural and morphological evaluations. These isolates are preserved at the National Institute of Biological Resources (NIBR) under the accession numbers NIBRFGC000510717 and NIBRFGC000510720, respectively.
The isolates were cultured on PDA for 10 days at 25°C for KNUF-23-232, and for 14 days at 25°C for KNUF-23-344 for the cultural and morphological characterization. Additionally, KNUF-23-232 was also cultured on oatmeal agar (OA; Difco, Detroit, MI, USA) for 10 days at 25°C. The cultures were maintained in darkness, and various characteristics were observed, including the size, color, and shape of the mycelium, as well as morphological features such as phialides, conidia, and the arrangement of conidia. Morphological properties were examined using a light microscope (BX-50; Olympus, Tokyo, Japan).
For molecular identification, total genomic DNA was extracted from strains KNUF-23-232 and KNUF-23-344 using the HiGene™ Genomic DNA Prep Kit for fungi (Biofact, Daejeon, Korea). The internal transcribed spacer (ITS) region, the large subunit of 28S rRNA (LSU), the second largest subunit of RNA polymerase II (RPB2), and translation elongation factor-1α (TEF1) were amplified for both strains. Additionally, the largest subunit of RNA polymerase II (RPB1) was specifically amplified for strain KNUF23-232. The primers used for amplifying the molecular phylogenetic markers were ITS1F/ITS4 for ITS, LROR/LR5 for LSU, RPB2-5F2/RPB2-7cR for RPB2, EF1-983F/EF1-2218R for TEF1, and cRPB1A/ RPB1Cr for RPB1 [8-16]. Successful amplification was confirmed by electrophoresis on 1.0% HP Agarose gels (BIOPURE, Cambridge, USA). The amplified products were purified using ExoSAP-IT (Thermo Fisher Scientific, Waltham, MA, USA) and subsequently submitted to Macrogen (Seoul, Korea) for sequencing.
The sequences obtained were analyzed for similarity using the Basic Local Alignment Search Tool (BLAST) within the National Center for Biotechnology Information (NCBI) database (Table 1). Phylogenetic trees were constructed from the concatenated sequences of the ITS regions, LSU, RPB1, RPB2, and TEF1, employing the neighbor-joining (NJ) method in MEGA 11 [17,18]. Evolutionary distance matrices for the NJ analysis were calculated using Kimura’s two-parameter model, with bootstrap values based on 1,000 replications [19].
Table 1. List of species used in this study and their corresponding GenBank accession numbers for phylogenetic analysis
| Species | Strain number | GenBank accession number | ||||
|---|---|---|---|---|---|---|
| ITS | LSU | RPB1 | RPB2 | TEF1 | ||
| Aciculosporium oplismeni | MAFF 246966 | LC571760 | LC571760 | – | LC572054 | LC572040 |
| Aciculosporium take | MAFF 241224 | LC571753 | LC571753 | – | LC572048 | LC572034 |
| Aciculosporium take | TNS-F-60465 | LC571755 | LC571756 | – | LC572049 | LC572035 |
| Balansia henningsiana | A.E.G. 96-27a | JN049815 | AY545727 | – | DQ522413 | AY489610 |
| Blackwellomyces aurantiacus | BCC 85060T | MT000692 | MT003028 | MK411600 | MT017819 | MK411598 |
| Blackwellomyces aurantiacus | BCC 85061 | MT000693 | MT003029 | MK411601 | MT017820 | MK411599 |
| Blackwellomyces cardinalis | OSC 93610 | JN049843 | AY184963 | EF469088 | EF469106 | EF469059 |
| Blackwellomyces calendulinus | BCC 68502T | MT000695 | MT003031 | MT017803 | MT017822 | MT017843 |
| Blackwellomyces calendulinus | BCC 68500 | MT000694 | MT003030 | MT017802 | MT017821 | MT017842 |
| Blackwellomyces calendulinus | KNUF-23-232 | PQ144579 | PQ144577 | PQ152981 | PQ148394 | PQ148396 |
| Blackwellomyces roseostromatus | BCC 91358T | MT000697 | MT003033 | MT017805 | MT017824 | MT017845 |
| Blackwellomyces roseostromatus | BCC 91359 | MT000698 | MT003034 | MT017806 | MT017825 | MT017846 |
| Blackwellomyces kaihuaensis | HMAS 285455T | OQ981975 | OQ981968 | OQ980409 | OQ980408 | OQ980401 |
| Blackwellomyces lateris | MFLU 18-0663T | MK086059 | MK086061 | MK084615 | MK079354 | MK069471 |
| Blackwellomyces minutus | BCC 88269T | MT000696 | MT003032 | MT017804 | MT017823 | MT017844 |
| Claviceps purpurea | GAM 12885 | U57669 | AF543789 | – | DQ522417 | AF543778 |
| Epichloë typhina | ATCC 56429 | JN049832 | U17396 | – | DQ522440 | AF543777 |
| Neoaraneomyces araneicola | DY101711T | MW730520 | MW730609 | – | MW753026 | MW753033 |
| Neoaraneomyces araneicola | DY101712 | MW730522 | MW730610 | – | MW753027 | MW753034 |
| Neoaraneomyces araneicola | KNUF-23-344 | PQ144580 | PQ144578 | – | PQ148395 | PQ148397 |
| P araneoaraneomyces sinensis | ZY 22.006T | OQ709254 | OQ709260 | – | OQ719621 | OQ719626 |
| P araneoaraneomyces sinensis | ZY 22.007 | OQ709255 | OQ709261 | – | OQ719622 | OQ719627 |
| P araneoaraneomyces sinensis | ZY 22.008 | OQ709256 | OQ709262 | – | OQ719623 | OQ719628 |
| Pleurocordyceps aurantiaca | MFLUCC 17-2113 | MG136916 | MG136910 | – | MG136870 | MG136875 |
| Pleurocordyceps marginaliradians | MFLU 17-1582 | MG136920 | MG136914 | – | MG271931 | MG136878 |
| Purpureocillium lilacinum | CBS 284.36T | MH855800 | FR775484 | EF468898 | EF468941 | EF468792 |
| Ustilaginoidea virens | MAFF 240421 | JQ349068 | JQ257011 | – | JQ257017 | JQ257024 |
ITS: Internal transcribed spacer regions; LSU: 28S rRNA large subunit; RPB1: the largest subunit of RNA polymerase II; RPB2: the second largest subunit of RNA polymerase II ; TEF1: translation elongation factor-1α.
T Type strain.
Strains used in this study are indicated in bold.
When cultured on PDA at 25℃ for 10 days, the colonies reached a diameter of 24‒26 mm. The obverse side appeared white and cream with some aerial hyphae, slightly wrinkled, and ciliate hyphae were observed at the edges along with reddish pigmentation (Fig. 1A). The reverse side of the colony exhibited a dark burgundy center, transitioning to scarlet toward the edges, with a white border (Fig. 1A). On OA, the colonies grew to a diameter of 22‒24 mm after 10 days at 25℃. The obverse side was white, with cottony aerial hyphae and a fringe-like edge, and the center of the colony was umbonate (Fig. 1B). The reverse side was light brown with no pigmentation (Fig. 1B). The phialides gradually tapered from a swollen basal portion to the apex, appearing solitary or in groups of two to three on each branch, arising from aerial hyphae (Fig. 1E-G). The phialides measured 12.5‒22.4 × 1.3‒2.5 μm (n=20). Conidia were hyaline, unicellular, elliptical to oblong-elliptical, with a smooth surface (Fig. 1H). The conidia measured 3.2‒5.3 × 1.6‒2.6 μm (n=50) and were produced at the tips of the hyphae in a Mariannaea-like conidial arrangement (Fig. 1C, 1D). A comparison of the morphological features of strain KNUF-23-232 with those of Blackwellomyces calendulinus BCC 68502T is provided in Table 2 [4]. While the reported B. calendulinus BCC 68502T exhibits both Mariannaea-like and Acremonium-like conidial arrangements, strain KNUF-23232 demonstrates only the Mariannaea-like arrangement. Other morphological and cultural characteristics observed indicate that strain KNUF-23-232 is most closely related to B. calendulinus BCC 68502T.
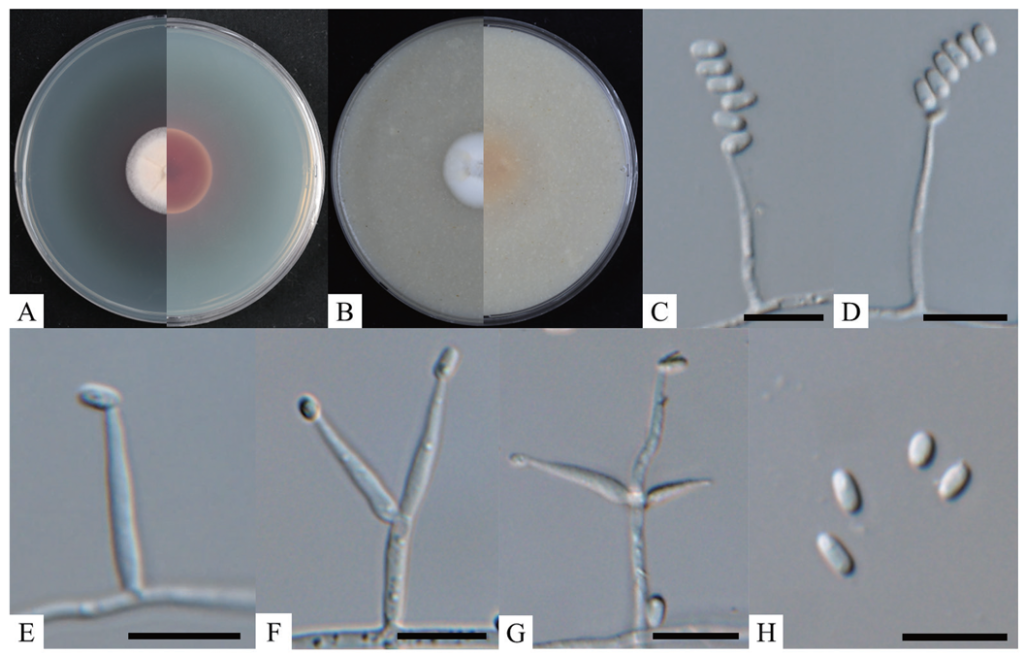
Fig. 1. Cultural and morphological characteristics of Blackwellomyces calendulinus KNUF-23-232. A, B: Obverse and reverse views of the colony at 25℃ after 10 days on potato dextrose agar (PDA) and oatmeal agar (OA), respectively; C, D: Mariannaea-like conidial arrangement; E-G: Phialides with 1-3 branches arising directly from aerial hyphae; H: Conidia. Scale bars = 10 μm.
Table 2. Morphological characteristics of Blackwellomyces calendulinus KNUF-23-232 compared with a previous report on B. calendulinus
| Characteristics | Blackwellomyces calendulinus KNUF-23-232a | Blackwellomyces calendulinus BCC 68502Tb | |
|---|---|---|---|
| Colony on PDA | Size (mm) | 24‒26 | 30 |
| Color | White to creamy, reverse center burgundy and margin scarlet, white fringe | White | |
| Shape | Circular, ciliate margin, slightly raised and wrinkled, cottony | Circular, ciliate margin, crateriform, cottony with high mycelium density | |
| Colony on OA | Size (mm) | 22‒24 | 25 |
| Color | White, reverse light brown | White | |
| Shape | Circular, ciliate margin, umbonate, cottony with high mycelium density | Circular, ciliate margin, umbonate, cottony with high mycelium density | |
| Conidia | Size (μm) | 3.2‒5.3 × 1.6‒2.6 | 3.5‒4.5 × 1.5‒2.0 |
| Shape | Hyaline, oblong-elliptical or ellipsoidal, smooth surface, one-celled | Hyaline, oblong-elliptical or ellipsoidal, smooth, one-celled | |
| Phialide | Size (μm) | 12.5‒22.4 × 1.3‒2.5 | 7.0‒12.0 × 1.0‒2.0 |
| Shape | Solitary or in whorls of two to three on each branch, gradually tapering from the base to the apex | Solitary or in whorls of two to five on each branch, gradually tapering from the base to the apex |
PDA: potato dextrose agar; OA: oatmeal agar.
T Type strain; a Fungal strain used in this paper; b Source of descriptions [4].
For the molecular identification of the isolated fungal strain KNUF-23-232, total genomic DNA was amplified to obtain sequences for the ITS region, LSU, RPB1, RPB2, and TEF1, yielding lengths of 500, 811, 637, 880, and 879 bp, respectively. The ITS regions showed 98.2% similarity with B. calendulinus BCC 68502T, whereas the LSU sequence displayed 99.4% similarity with B. calendulinus BCC 85060T. The RPB1 sequence demonstrated 98.7% with B. calendulinus BCC 68502T and 98.0% similarity with B. lateris MFLU 18-0663T. The RPB2 sequence revealed 99.8% with B. calendulinus BCC 68502T and 97.0% with B. roseostromatus BCC 91358T. The TEF1 sequences showed 99.9% similarity with B. calendulinus BCC 68502T. A phylogenetic tree was constructed using the NJ method based on the concatenated sequences of the ITS region, LSU, RPB1, RPB2, and TEF1 (Fig. 2). In this phylogenetic tree, strain KNUF23-232 clustered closely with B. calendulinus BCC 68502T. Based on the cultural, morphological, and phylogenetic analyses, strain KNUF-23-232 was identified as B. calendulinus.
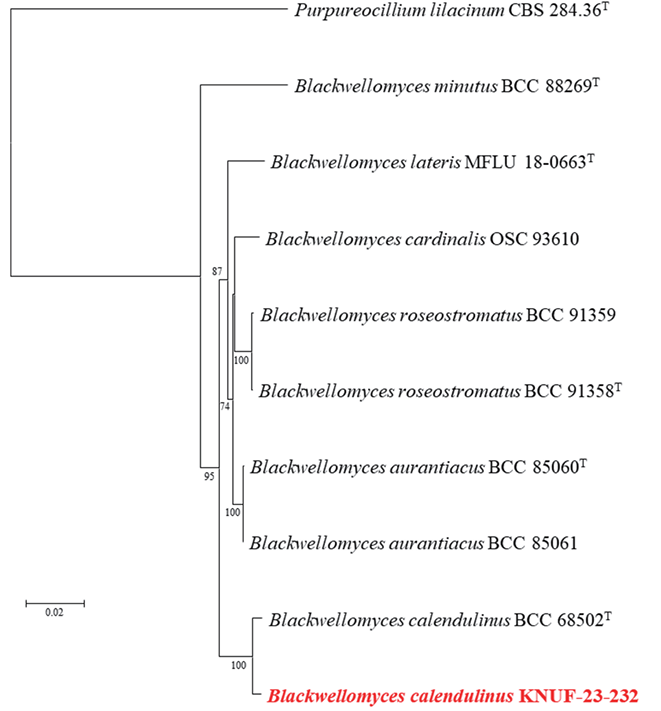
Fig. 2. Neighbor-joining phylogenetic analysis of KNUF-23-232 based on concatenated sequence data from the internal transcribed spacer (ITS) regions, large subunit of 28S rRNA (LSU), the largest subunit of RNA polymerase II (RPB1), the second largest subunit of RNA polymerase II (RPB2) and translation elongation factor-1α (TEF1), illustrating the phylogenetic position of the closest species within the genus Blackwellomyces. Bootstrap values greater than 70% (based on 1,000 replications) are indicated at branch points. Purpureocillium lilacinum CBS 284.36T was used as an outgroup. The strain isolated in this study is highlighted in bold red. Bar=0.02 substitutions per nucleotide position.
The genus Blackwellomyces comprises eight species reported globally, including B. cardinalis, B. pseudomilitaris, B. aurantiacus, B. roseostromatus, B. lateris, and B. kaihuaensis, which have been isolated from lepidopteran larvae, and B. calendulinus and B. minutus from coleopteran larvae [20]. While all species within this genus have been isolated from insect hosts, there is a notable lack of studies assessing their pathogenicity or investigating their potential as insect pathogens. Genetic analysis of B. cardinalis NBRC 103832 has revealed that the red pigment produced in the culture medium is oosporein, a secondary metabolite known to inhibit bacterial growth [21]. This finding suggests the potential for entomopathogenic characteristics within the genus Blackwellomyces. Although no studies have specifically examined the secondary metabolites or entomopathogenicity of B. calendulinus, it is posited that this species may possess significant potential as an entomopathogenic fungus, warranting further investigation.
When cultured on PDA at 25℃ for 14 days, the colonies reached a diameter of 24‒28 mm. The obverse side appeared white to light yellow, with an umbonate shape and wrinkles radiating from the center to the edges, which were irregularly undulate (Fig. 3A). The reverse side exhibited similar characteristics, with wrinkles extending from the center to the edges and a yellowish color (Fig. 3A). The conidiophores were observed to be mononematous, arising laterally from the aerial hyphae. The phialides were observed to be cylindrical to ellipsoidal, tapering from the base towards the apex (Fig. 3B), with dimensions ranging from 12.0‒31.2 × 1.0‒1.9 μm (n=20). The conidia, produced at the tip of the phialides, were cateniferous, resembling those of Paecilomyces (Fig. 3C). These conidia were hyaline, fusiform to ellipsoidal or semiorbicular, unicellular, and measured 2.8‒4.8 × 1.1‒2.0 μm in length (n=50) (Fig. 3D). A comparative analysis of the morphological features of strain KNUF-23-344 with Neoaraneomyces araneicola DY101711T and Paraneoaraneomyces sinensis ZY 22.006T is presented in Table 3, indicating that strain KNUF-23-344 is closely related to N. araneicola DY101711T.
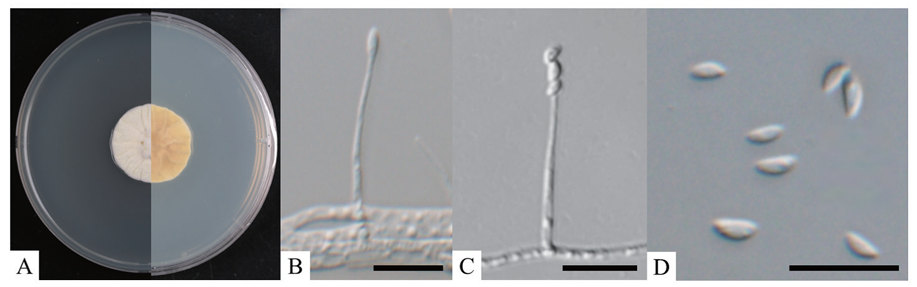
Fig. 3. Cultural and morphological characteristics of Neoaraneomyces araneicola KNUF-23-344. A: Obverse and reverse views of the colony at 25℃ after 14 days on potato dextrose agar (PDA); B: Phialide; C: Phialide with conidia in chain; D: Conidia. Scale bars=10 μm.
Table 3. Morphological characteristics of Neoaraneomyces araneicola KNUF-23-344 compared with a previous report on N. araneicola
| Characteristics | Neoaraneomyces araneicola KNUF-23-344a | Neoaraneomyces araneicola DY101711Tb | |
|---|---|---|---|
| Colony on PDA | Size (mm) | 24‒28 | 30‒32 |
| Color | White to light yellow, reverse yellowish | White to pale grey, reverse yellowish | |
| Shape | Irregular and undulate margin, umbonate, wrinkled | Irregular and undulate margin, umbonate, wrinkled, consisting of a basal felt | |
| Phialide | Size (μm) | 12.0‒31.2 × 1.0‒1.9 | 8.9‒23.8 × 1.1‒1.6 |
| Shape | Mononematous, arising from the lateral, aerial hyphae | Mononematous, arising from the lateral, aerial hyphae | |
| Conidia | Size (μm) | 2.8‒4.8 × 1.1‒2.0 | 2.9‒4.4 × 1.3‒2.0 |
| Shape | Hyaline, fusiform to ellipsoidal or semiorbicular, cateniferous, one-celled | Cateniferous, hyaline, fusiform to ellipsoidal, onecelled |
PDA: potato dextrose agar; N/A: not available.
T Type strain; a Fungal strain used in this paper; b Source of descriptions [5].
For the molecular identification of the isolated fungal strain, the total genomic DNA of KNUF-23-344 was amplified to obtain the ITS region, LSU, RPB2, and TEF1 (343, 416, 594, and 780 bp, respectively). The ITS regions exhibited a high 99.5% similarity to N. araneicola DY101711T, whereas the LSU sequences demonstrated 100% similarity with both N. araneicola DY101711T and DY101712. The RPB2 sequences exhibited a 99.0% similarity with N. araneicola DY101711T and a 98.7% similarity with N. araneicola DY101712. The TEF1 sequences showed a 99.8% similarity with both N. araneicola DY101711T and DY101712. A phylogenetic tree was constructed using the NJ method with the concatenated sequences of the ITS regions, LSU, RPB2, and TEF1 (Fig. 4). Based on the NJ phylogenetic tree, strain KNUF-23-344 clustered closely with N. araneicola DY101711T and DY101712. The cultural, morphological, and phylogenetic analyses collectively identified strain KNUF-23-344 as N. araneicola.
The genus Neoaraneomyces was established by Chen et al. in 2022, with only one species, N. araneicola, reported, making it a highly rare taxon [5]. Among the 13 recognized genera of araneogenous fungi, only the genus Gibellula is documented to exclusively parasitize spiders. However, N. araneicola exhibits distinct characteristics that differentiate it from Gibellula, particularly in its conidial structure, which resembles that of Paecilomyces, with chain-like conidia observed at the apex of the hyphae. [5,22]. To date, no studies have addressed the pathogenicity of N. araneicola. Therefore, the KNUF-23-344 strain isolated in this study warrants further research on the pathogenicity of N. araneicola and its metabolites, with potential future applications as an acaricide in the future. To the best of our knowledge, this is the first report of these two species in Korea. The unreported species, B. calendulinus and N. araneicola, isolated in this study, will contribute to the understanding and conservation of domestic biodiversity. Additionally, they provide valuable data that may enhance the potential utilization of their entomopathogenic properties.

Fig. 4. Phylogenetic analysis of KNUF-23-344 using the neighbor-joining method based on concatenated sequence data from the internal transcribed spacer (ITS) regions, large subunit of 28S rRNA (LSU), the second largest subunit of RNA polymerase II (RPB2) and translation elongation factor-1α (TEF1), showing the phylogenetic position of the closest genus within the family Clavicipitaceae. Bootstrap values greater than 80% (based on 1,000 replications) are indicated at branch points. Pleurocordyceps aurantiaca MFLUCC 17-2113 was used as an outgroup. The strain isolated in this study is highlighted in bold red. Bar=0.02 substitutions per nucleotide position.
The authors declare no conflict of interest.
This research was supported by a grant from the National Institute of Biological Resources (NIBR), funded by the Ministry of Environment (MOE) of the Republic of Korea (NIBR202304104).
1. Ma M, Luo J, Li C, Eleftherianos I, Zhang W, Xu L. A life-and-death struggle: interaction of insects with entomopathogenic fungi across various infection stages. Front Immunol 2024;14:1-13. [DOI]
2. Zhang W, Chen X, Eleftherianos I, Mohamed A, Bastin A, Keyhani NO. Cross-talk between immunity and behavior: insights from entomopathogenic fungi and their insect hosts. FEMS Microbiol Rev 2024;48:1-14. [DOI]
3. Kepler RM, Luangsa-Ard JJ, Hywel-Jones NL, Quandt CA, Sung GH, Rehner SA, Aime MC, Henkel TW, Sanjuan T, Zare R. A phylogenetically-based nomenclature for Cordycipitaceae (Hypocreales). IMA Fungus 2017;8:335-53. [DOI]
4. Mongkolsamrit S, Noisripoom W, Tasanathai K, Khonsanit A, Thanakitpipattana D, Himaman W, Kobmoo N, Luangsa-Ard JJ. Molecular phylogeny and morphology reveal cryptic species in Blackwellomyces and Cordyceps (Cordycipitaceae) from Thailand. Mycol Prog 2020;19:957-83. [DOI]
5. Chen WH, Liang JD, Ren XX, Zhao JH, Han YF, Liang ZQ. Phylogenetic, ecological and morphological characteristics reveal two new spider-associated genera in Clavicipitaceae. MycoKeys 2022;91:49-66. [DOI]
6. Nyffeler M, Hywel-Jones N. Diversity of spider families parasitized by fungal pathogens: a global review. bioRxiv 2023:1-86. [DOI]
7. Das K, You YH, Lee SY, Jung HY. A new species of Thelonectria and a new record of Cephalotrichum hinnuleum from Gunwi and Ulleungdo in Korea. Mycobiology 2020;48:341-50. [DOI]
8. Gardes M, Bruns TD. ITS primers with enhanced specificity for basidiomycetes‐application to the identification of mycorrhizae and rusts. Mol Ecol 1993;2:113-8. [DOI]
9. White TJ, Bruns TD, Lee SB, Taylor JW. Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics. In: Innis MA, Gelfand DH, Sninsky JJ, editors. PCR protocols: a guide to methods and applications. San Diego: Academic Press; 1990. p. 315-22. [DOI]
10. Rehner SA, Samuels GJ. Taxonomy and phylogeny of Gliocladium analysed from nuclear large subunit ribosomal DNA sequences. Mycol Res 1994;98:625-34. [DOI]
11. Vilgalys R, Hester M. Rapid genetic identification and mapping of enzymatically amplified ribosomal DNA from several Cryptococcus species. J Bacteriol 1990;172:4238-46. [DOI]
12. Sung GH, Sung JM, Hywel-Jones NL, Spatafora JW. A multi-gene phylogeny of Clavicipitaceae (Ascomycota, Fungi): Identification of localized incongruence using a combinational bootstrap approach. Mol Phylogenet Evol 2007;44:1204-23. [DOI]
13. Liu YJ, Whelen S, Hall BD. Phylogenetic relationships among ascomycetes: evidence from an RNA polymerse II subunit. Mol Biol Evol 1999;16:1799-808. [DOI]
14. Rehner SA, Buckley E. A Beauveria phylogeny inferred from nuclear ITS and EF1-α sequences: Evidence for cryptic diversification and links to Cordyceps teleomorphs. Mycologia 2005;97:84-98. [DOI]
15. Castlebury LA, Rossman AY, Sung GH, Hyten AS, Spatafora JW. Multigene phylogeny reveals new lineage for Stachybotrys chartarum, the indoor air fungus. Mycol Res 2004;108:864-72. [DOI]
16. Matheny PB, Liu YJ, Ammirati JF, Hall BD. Using RPB1 sequences to improve phylogenetic inference among mushrooms (Inocybe, Agaricales). Am J Bot 2002;89:688-98. [DOI]
17. Saitou N, Nei M. The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol Biol Evol 1987;4:406-25.
18. Tamura K, Stecher G, Kumar S. MEGA11: molecular evolutionary genetics analysis version 11. Mol Biol Evol 2021;38:3022-7. [DOI]
19. Kimura M. A simple method for estimating evolutionary rates of base substitutions through comparative studies of nucleotide sequences. J Mol Evol 1980;16:111-20. [DOI]
20. Li Y, Zhao XC, Wu LX, Wang Y, Xu A, Lin WF. Blackwellomyces kaihuaensis and Metarhizium putuoense (Hypocreales), two new entomogenous fungi from subtropical forests in Zhejiang Province, Eastern China. Forests 2023;14:1-19. [DOI]
21. Nakamura Y, Nguyen NH, Yoshinari T, Hachisu M, Nguyen PT, Shimizu K. Identification of the oosporein biosynthesis gene cluster in an entomopathogenic fungus Blackwellomyces cardinalis. Mycoscience 2024;65:96-104. [DOI]
22. Shrestha B, Kubátová A, Tanaka E, Oh J, Yoon DH, Sung JM, Sung GH. Spider-pathogenic fungi within Hypocreales (Ascomycota): their current nomenclature, diversity, and distribution. Mycol Prog 2019;18:983-1003. [DOI]