Hee-Young Jung1,2,*, Soo-Min Hong1, Seong-Keun Lim1, Song-Woon Nam1, Leonid N. Ten2, Won-Kwon Jung3, and Seung-Yeol Lee1,2
1Department of Plant Medicine, Kyungpook National University, Daegu 41566, Korea
2Institute of Plant Medicine, Kyungpook National University, Daegu 41566, Korea
3Gyeongsangbuk-do Agricultural Research and Extension Services, Daegu 41404, Korea
*Correspondence to heeyoung@knu.ac.kr
Korean Journal of Mycology (Kor J Mycol) 2024 September, Volume 52, Issue 3, pages 223-232.
https://doi.org/10.4489/kjm.520308
Received on May 20, 2024, Revised on September 25, 2024, Accepted on September 25, 2024, Published on Sep 30, 2024.
Copyright © The Korean Society of Mycology.
This is an Open Access article which is freely available under the Creative Commons Attribution-NonCommercial 4.0 International License (CC BY-NC) (https://creativecommons.org/licenses/by-nc/4.0/).
Strain KNUF-MG-01 was isolated from a mango tree exhibiting witch’s broom symptoms in Gimhae-si and Gunwi-gun, South Korea. After 7 days of incubation on potato dextrose agar (PDA), the colonies displayed white to rose mycelia and reached dimensions of 79.6‒80.1 mm. Microconidia were abundant, mostly straight or slightly curved, non or one septate, measuring 4.6‒7.5‒13.9 × 2.0‒3.1‒6.4 μm. Macroconidia, characterized by being straight or slightly curved, slender, and 2 to 3 septate, measured 21.4‒36.7 × 3.5‒3.8 μm. Multilocus sequence analysis, using a combined dataset of internal transcribed spacer (ITS) regions, translation elongation factor-1α (TEF1-α), and β-tubulin (β-TUB) gene sequences, identified KNUF-MG-01 as a strain of Fusarium mangiferae. To our knowledge, this is the first report of F. mangiferae in Korea.
Fusarium mangiferae, mango, morphology, phylogeny, unreported species
Mango (Mangifera indica L.) is a key global fruit crop, with a production of approximately 60 million tons in 2022 [1]. Despite being a relatively minor contributor to global mango production, Korea cultivated approximately 407 tons in 2021 over a total land area of 40 ha, according to Statistics Korea (KOSTAT) [2]. Mango cultivation worldwide faces a significant threat from mango malformation disease (MMD), which adversely affects the growth and productivity of this essential fruit crop [3,4]. MMD was first documented in India in 1891 [5] and subsequently spread to various regions, including Asia, South Africa, the USA, and several other countries [6]. The genus Fusarium, which causes MMD, includes more than 400 species cataloged in the Mycobank (https://www.mycobank.org) and Index Fungorum (http://www. indexfungorum.org) databases. The genus Fusarium primarily targets potatoes, peas, beans, and members of the Cucurbitaceae family. Plant-pathogenic Fusarium strains also cause fruit rot in guavas, bananas, papayas, and other tropical fruits [7]. Research in India has shown that the primary cause of floral and vegetative malformation in mangoes is attributed to the presence and influence of F. moniliforme [8]. In 1974, strains of F. moniliforme var. subglutinans, isolated from vegetative shoots and floral tissue, were used to induce the floral form of MMD [9]. Subsequently, these strains were reclassified as F. subglutinans [10]. Artificial inoculation experiments reveal that certain strains of F. subglutinans from Egypt, Oman, Florida, Israel, Malaysia, and South Africa can induce MMD [11]. In 2002, a new Fusarium species, F. mangiferae, consisting of 29 phylogenetically related strains, was identified [12]. It has been detected in China, Egypt, India, Israel, Malaysia, Oman, South Africa, Spain, Sri Lanka, and the USA, becoming the predominant causal agent of MMD worldwide [13]. It is noteworthy that F. mangiferae has been determined to be conspecific with strains previously identified as F. subglutinans, which were reported as the causative agent of malformation in mango-growing regions worldwide [14]. In 2016, a specific primer pair was developed to rapidly detect F. mangiferae, thereby facilitating MMD detection caused by this pathogen [15]. Besides F. mangiferae, several other species from the F. fujikuroi species complex (FFSC), including F. mexicanum, F. proliferatum, F. sterilihyphosum, and F. tupiense, have been identified in association with MMD [16]. Recent research has shown a distinct pattern in mango buds infected with F. mangiferae. It is characterized by significantly reduced levels of gibberellic acid (GA) and markedly increased levels of auxin [17]. Furthermore, the invasive action of F. mangiferae in mango buds disrupts the nutrient allocation system and endogenous hormone balance by utilizing carbohydrates or newly synthesized GAs, as well as externally applied GAs [18]. This compelling evidence establishes F. mangiferae as a key causal agent of MMD, especially through the hormonal profiling of infected mango buds by F. mangiferae concerning mango malformation. In this study, Fusarium species designated as KNUF-MG-01, KNUF-MG-02, and KNUFMG-03 were isolated from MMD-like symptoms. To identify the isolated Fusarium species, cultural, morphological, and molecular analyses were conducted in this study. Here, the findings are documented and reported below.
The fungal strains used in this study were isolated from symptoms of a witch’s broom-like appearance observed on mango trees located in Gimhae-si (35°15’39.3″N 128°45’46.7″E) and Gunwi-gun (36°06’19.1″N 128°37’40.1″E), South Korea. MMD occurrences were observed across mango trees aged 2‒5 years (Fig. 1). The small pieces of the diseased plant tissues were sterilized using 70% ethanol and 1% sodium hypochlorite, followed by three washes of sterile water. The tissues were then placed on potato dextrose agar (PDA; Difco, Detroit, MI, USA) and incubated at 25℃. After 2 days, free from any bacterial or fungal contamination, the hyphal tips from the developing fungus were transferred to a new PDA plate to obtain pure fungal isolates. Strains KNUF-MG-01, KNUF-MG-02, and KNUF-MG-03 were selected from numerous fungal strains for additional molecular and phylogenetic analyses. The fungal strains were preserved at −80℃ in 20% glycerol stock.

Fig. 1. Typical symptoms of mango malformation disease caused by Fusarium mangiferae. A-C: Development of multiple stiff leaves. D: Vegetative malformation in mango featuring branches and flowers with deformed shoots, short internodes, and stiff leaves. E: Development of multiple stiff leaves. F: Leaves turn brown-black and dry as they age. G: Symptoms of leaf spot on mango (red arrows). A-C: Five-year-old mango trees collected from Gimhae-si, South Korea; D-G: Two-year-old mango trees collected from Gunwi-gun, South Korea.
To examine the cultural and morphological characteristics, the isolated fungal strain KNUF-MG-01 was cultured on PDA and synthetic nutrient agar (SNA) [19]. The color of the colony, shape, and growth rate characteristics were observed after 7 days. Macroconidia production was observed by growing strain KNUF-MG-01 on SNA medium for 14 days at 25℃ and examining it using a BX-50 microscope (Olympus, Tokyo, Japan).
Total genomic DNA of the strains KNUF-MG-01, KNUF-MG-02, and KNUF-MG-03 were extracted using a commercial extraction kit, HiGene Genomic DNA Prep Kit (Biofact, Daejeon, Korea), according to the manufacturer’s instructions. Molecular identification was conducted by analyzing sequences of the internal transcribed spacer (ITS) regions and large subunit (LSU) rRNA, β-tubulin (TUB2), and RNA polymerase II subunit (RPB2) genes, which were amplified separately using the primer pairs ITS1F/ ITS4, T1/T2, and EF1/EF2, respectively [20-24]. The quality of PCR products was analyzed by 0.7% agarose gel electrophoresis and stained with ethidium bromide. PCR products were purified using ExoSAP-IT PCR Product Cleaning Reagent (Thermo Fisher Scientific, Waltham, MA, USA) and sequenced by Bioneer (Daejeon, South Korea).
The gene sequences were organized using the SeqMan software version 2 (DNASTAR, Madison, WI, USA). The editing of gaps and terminal ends in the alignment was performed using BioEdit version 5.0.6. to ensure the accuracy and completeness of the gene sequences. To identify the percentage of sequence similarity with other fungal species, the gene sequences were compared with those in the NCBI database using a BLASTn search. Additionally, Fusarium sequences were retrieved from NCBI’s GenBank to identify phylogenetic relationships (Table 1). The phylogenetic tree was constructed based on the concatenated sequences of the ITS regions, TEF1-α, and β-TUB genes with MEGA 7 software, employing the neighbor-joining method and 1,000 bootstrap replicates [25]. Genetic divergence between the species was assessed using Kimura’s two-parameter model [26].
Table 1. Fusarium strains and corresponding Genbank accession numbers used in phylogenetic analyses
| Species | Strain | GenBank accession numbers | References | ||
|---|---|---|---|---|---|
| ITS | TEF1-α | β-TUB | |||
| Fusarium chlamydosporum | BCCM/IHEM:24710 | KJ125534 | KJ126126 | KJ125830 | [27] |
| Fusarium circinatum | CBS 405.97 | MH862654 | KM231943 | KM232080 | [27] |
| Fusarium equiseti | UP-PA002 | MH521295 | MH521297 | MH521296 | [27] |
| Fusarium fujikuroi | CBS 257.52 | MH857023 | MW402119 | KU603885 | [27] |
| Fusarium incarnatum | JS21 | MT889974 | MT895846 | MT895843 | [27] |
| Fusarium mangiferae | CBS 119853 | MH863065 | MN534016 | MN534140 | [27] |
| Fusarium mangiferae | GZ14-G-2 | MT358793 | MK524418 | MK347269 | [27] |
| Fusarium mangiferae | NRRL 25226 | U61691 | AF160281 | U61561 | [27] |
| Fusarium mangiferae | KNUF-MG-01 | LC802131 | LC802133 | LC802132 | This study |
| Fusarium oxysporum | B2 | MZ060273 | MN754062 | MN754078 | [27] |
| Fusarium proliferatum | M259 | KJ467095 | KJ555080 | KJ544176 | [27] |
| Fusarium proliferatum | NRRL 22944 | U34558 | AF160280 | U34416 | [27] |
| Fusarium proliferatum | YB2-1 | MT358796 | MK524423 | MK347274 | [27] |
| Fusarium solani | LC13843 | MW016729 | MW620190 | MW534069 | [27] |
| Fusarium solani | LC3717 | MW016736 | MW620197 | MW534074 | [27] |
| Fusarium subglutinans | CBS 136481 | KR071625 | KU711692 | KU603893 | [27] |
| Fusarium thapsinum | CBS 130176 | KR071692 | MW402022 | MW402222 | [27] |
| Fusarium verticillioides | E52 | KJ467098 | KJ555083 | KJ544181 | [27] |
| Fusarium virguliforme | NRRL 22825 | GU170655 | EF408437 | EF408472 | [27] |
ITS: internal transcribed spacer; TEF1-α: partial translation elongation factor-1α; β-TUB: β-tubulin.
The isolated strain is shown in bold.
The strains KNUF-MG-01, KNUF-MG-02, and KNUF-MG-03 were found to be morphologically identical, and they clustered together for molecular phylogeny. Since they were identical, only the cultural and morphological characteristics of strain KNUF-MG-01 were described in this study.
The colonies developed white to pink mycelia, reaching a diameter of 79.6‒80.1 mm on the PDA after 7 days (Fig. 2A and B). On SNA, the fungus formed a whitish, cottony colony with aerial mycelia and f loccose, covering the Petri dish after 7 days (Fig. 2C and D). The macroconidia were slightly bent, sicklelike, measured 21.4‒36.7 × 3.5‒3.8 μm on average, mostly 2‒3 septate, with rounded apical cells (Fig. 3A). Microconidia were predominantly oval to ellipsoidal, non-septate or one-septate, and abundantly produced, with an average diameter ranging 4.6‒7.5‒13.9 × 2.0‒3.1‒6.4 μm (Fig. 3B) from elongated phialides (Fig. 3C). These cultural and morphological features of strain KNUF-MG-01 was closely comparable to those of F. mangiferae [12], with a slight difference observed in the range of number of macroconidia septa (Table 2). In contrast, KNUF-MG-01 differed from its relative, F. fujikuroi, in exhibiting larger microconidia (4.6‒7.5‒13.9 × 2.0‒3.1‒6.4 μm vs. 5‒13 × 2.1‒4.7 μm), smaller macroconidia (21.4‒36.7 × 3.5‒3.8 μm vs. 22‒59 × 2.5‒4.7 μm), and typically having a relatively lower number of macroconidia septa (2‒3 vs. 3‒5). Cultural characteristics, along with the size and shape of microconidia and macroconidia, distinguish strain KNUF-MG-01 from another phylogenetically closely related species, F. proliferatum (Table 2). KNUFMG-01 exhibited a larger colony size (79.6‒80.1 mm vs. 69‒71 mm) on PDA after 7 days of growth at 25℃, displayed a different color on the reverse side of the colony, and produced oval to ellipsoidal microconidia with significantly shorter macroconidia (21.4‒36.7 μm) compared to those of F. proliferatum (19‒59 μm).
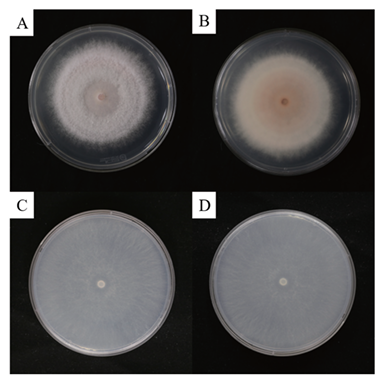
Fig. 2. Cultural characteristics of Fusarium mangiferae KNUF-MG-01. A: Colony on potato dextrose agar (PDA) after 7 days of growth at 25℃. B: The reverse side of the colony on PDA displays a white-to-pink pigment. C: Colony on SNA after 7 days of growth at 25℃. D: The reverse side of the colony on synthetic nutrient agar (SNA) shows a cottony texture with a whitish pigment.
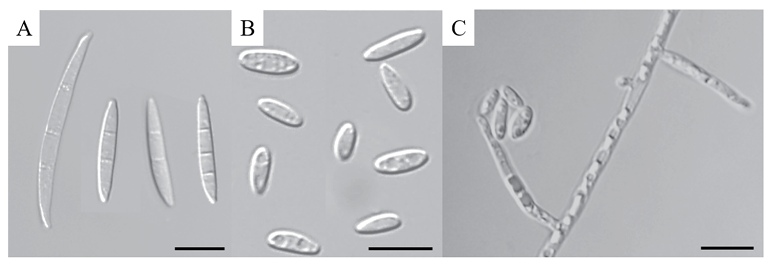
Fig. 3. Observed morphological characteristics of Fusarium mangiferae KNUF-MG-01. A: Macroconidia observed on synthetic nutrient agar (SNA) media. B: Microconidia, predominantly 0-1 septate and oval in shape. C: Phialides from which microconidia are produced and accumulated. Scale bars = 10 µm.
Table 2. Morphological comparison of strain KNUF-MG-01 with phylogenetically related Fusarium species
| Characteristics | Fusarium mangiferae KNUF-MG-01a | Fusarium mangiferae PREM 57299b | Fusarium fujikuroi CBS 221.76c | Fusarium proliferatum CBS 480.96d | |
|---|---|---|---|---|---|
| Colony | Color | Center white to pink, reverse rosy buff, outer region whitish | Center white, sometimes reverse rosy buff to dark purple | Whitish to pale pink | Surface floccose, white to pale pink. Reverse with pale greyish ruby center fading into a greyish rose, becoming dark violet to blue at center with age. |
| Size (mm) | Reaching 79.6‒80.1 mm on PDA after 7 days | 3.40 mm/day at 25℃ on PDA | 7‒8 cm in 10 days at 25℃ on PDA | 69‒71 mm AFter 7 days at 25℃ on PDA | |
| Aerial mycelium | Floccose aerial mycelium | Floccose aerial mycelium | Abundant, fluffy, l anuginose, not felt-like, whitish to pale pink | Abundant, pannose, later tufts of white patch-mutants appear, white to pinkish buff, in older cultures pale to dark vinaceous | |
| Microconidia | Mostly oval to ellipsoidal, non or 1 septate; 4.55‒7.51‒13.93 × 1.95‒3.11‒6.35 μm | Variable in shape, oval to allantoid conidia occurring o ccasionally; 0 septate or 1 septate; 0 septate 4.3‒9.0‒14.4 × 1.7‒2.4‒3.3 μm | Clavate with a flattened base, mostly 1-celled, rarely 1-septate; 0 septate mostly 7‒10 × 2.5‒3.3 μm (5‒13 × 2.1‒4.7 μm) | Primarily clavate with a flattened base, usually 1-celled, exceptionally 1-septate; 0 septate mostly 7‒11 × 4.7‒7.7 μm (5‒15 × 3.3‒12.4 μm) on pyriform; in case of clavate, the 0 septate mostly 7‒9 × 2.2‒3.2 μm (3‒16 × 1.2‒5.3 μm) | |
| Macroconidia | Slightly bent, sickle-like, 21.37‒36.72 × 3.46‒3.77 μm on average, mostly 2‒3 septate, and with rounded apical cells | Long and slender; usually 3 –5-septate, 43 .1‒51.8‒61.4 × 1.9‒2.3‒3.4 μm | Usually 3 to 5 septate; 3 septate mostly 33‒48 × 3.0‒3.9 μm (22‒59 × 2.5‒4.7 μm); 5 septate mostly 50‒59 × 3.6‒3.9 μm (43‒65 × 3.3‒4.4 μm) | Falcate but rather straight with a distinct foot cell, usually 3-septate or 5-septate, 3-septate mostly 30‒46 × 3.3‒4.1 μm (19‒59 × 2.6‒5.0 μm); 5-septate mostly 47‒58 × 3.4‒4.4 μm (39‒79 × 3.0‒5.0 μm) | |
| Pigments on PDA | White to pink | Light-to dark-purple | Initially buff, later rust to brick | Buff, grey-lilac, brick to brown-vinaceous |
PDA: potato dextrose agar
a Fungal strain used in this paper; b Source of descriptions [12]; c Source of description [27]; d Source of description [27,28].
The sequence length of the ITS region of strain KNUF-MG-01 was 575 bp. The ITS regions of KNUF-MG-01 showed 100% and 99.8% similarities with F. proliferatum DSM 106835 (GenBank no. MH055399) and F. fujikuroi NBRC 30337 (GenBank no. AB237662), respectively. This analysis demonstrated that relying solely on a comparative examination of the ITS region sequence is insufficient for accurate species-level classification of strain KNUF-MG-01. Consequently, sequences from the TEF1-α and β-TUB genes were utilized for further phylogenetic analysis. The β-TUB gene sequence (572 bp) of strain KNUF-MG-01 was 100% and 99.9% identical to that of F. mangiferae GD34 (GenBank no. MK347264) and F. proliferatum CBS 480.96 (GenBank no. MN534129), respectively. A BLAST search for the TEF1-α gene sequence (965 bp) of the isolate revealed identities of 100, 100, 99.9, and 99.7% with closely related F. mangiferae GX13-3 (Genbank no. MK524414), F. lumajangense LC13650 (Genbank no. MW580501), F. proliferatum Lido02 (Genbank no. OM634654), and F. concentricum LC13620 (Genbank no. MW580456), respectively. Overall, the examination of TEF1-α and β-TUB gene sequences did not yield a precise identification of the new strain at the species level, similar to the findings for the ITS regions. In the case of Fusarium species, various gene combinations are commonly employed for phylogenetic analysis. In particular, the combined ITS regions, TEF1-α, and β-TUB gene sequences allowed precise identification of F. thapsinum that caused sorghum leaf spot disease [27]. The same approach for phylogenetic analysis was used in our study. The topology of the neighbor-joining phylogenetic tree, based on the concatenated sequences of ITS regions, TEF1-α and β-TUB genes, indicated that strain KNUF-MG-01 clustered together with F. mangiferae strains (Fig. 4), confirming its closest relationship at the species level. Thus, based on morphological and phylogenetic analyses, strain KNUF-MG-01 was identified as F. mangiferae, a newly described fungus in Korea.
In 2020, the pathogenicity of F. mangiferae towards mango fruits was confirmed through identification and molecular characterization [28]. Furthermore, the isolation, identification, and artificial inoculation of mango buds with F. mangiferae (IARI isolate KF060921) under controlled conditions led to symptoms of mango malformation [16]. In 2016, inoculation with F. mangiferae resulted in short internodes with swollen buds and small scale-like leafy structures, causing the arrested growth of shootlets and the continuous emergence of similar malformed bunches [29]. Although pathogenicity tests were not conducted on strains KNUF-MG-01, KNUF-MG-02, and KNUF-MG-03 reported in this study, as previously mentioned, various research of plant diseases caused by F. mangiferae have been reported worldwide. Therefore, further research, such as pathogenicity tests, should be conducted to verify the pathogenicity of strains KNUFMG-01, KNUF-MG-02, and KNUF-MG-03. Additionally, the identification of F. mangiferae, a previously unreported species in Korea, is expected to expand knowledge of its ecological distribution in the Korean environment condition.
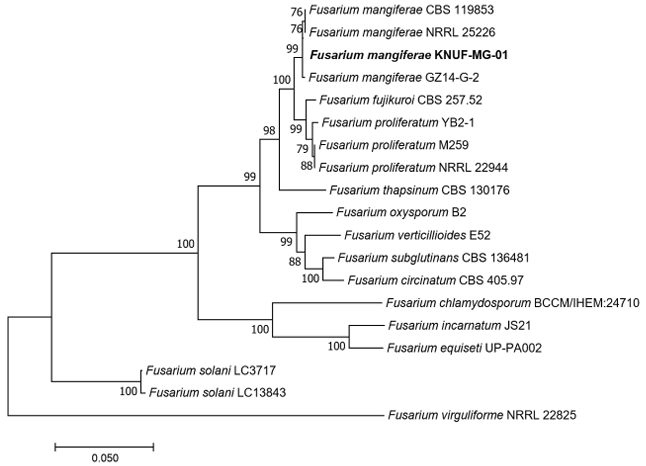
Fig. 4. Neighbor-joining phylogenetic tree showing the relationships between Fusarium mangiferae KNUF-MG-01 and other Fusarium species, based on concatenated sequences of the internal transcribed spacer (ITS) regions, translation elongation factor-1α (TEF1-α) and β-tubulin (β-TUB) genes. Bootstrap values greater than 70% (based on 1,000 replications) are shown at the branch points. The strain isolated in this study is highlighted in bold. The scale bar represents 0.050 substitutions per nucleotide position.
The authors declare that they have no potential conflicts of interest.
This work was supported by the Korea Institute of Planning and Evaluation for Technology in Food, Agriculture and Forestry (IPET) through the High Value-added Food Technology Development Program, funded by the Ministry of Agriculture, Food and Rural Affairs (MAFRA) (RS-2024-00403514).
1. Food and Agriculture Organization of the United Nations. Mangoes, guavas and mangosteens production indices (Production) [Internet]. Rome: FAOSTAT; 2022 [cited 2024 Sep 23]. Available from http://faostat.fao.org.
2. Statistics Korea. Fruit production [Internet]. Daejeon: KOSTAT; 2022 [cited 2024 Sep 23]. Available from https://kostat.go.kr.
3. Kumar J, Singh US, Beniwal SPS. Mango malformation: one hundred years of research. Annu Rev Phytopathol 1993;31:217-32. [DOI]
4. Ploetz RC, Freeman S. Foliar, floral and soilborne diseases. In: Litz RE, editor. The Mango: Botany Production and Uses, 2nd edition. Wallingford: CAB International; 2009. p. 231-302. [DOI]
5. Ploetz RC. Malformation: A unique and important disease of mango, Mangifera indica L. In: Summerell BA, Leslie JF, Backhouse D, Bryden WL, Burgess LW, editors. Fusarium: Paul E. Nelson Memorial Symposium. St. Paul, MN: The American Phytopathological Society; 2001. p. 233-47.
6. Katoch P, Katoch A, Dangi B. An overview on mango malformation and the potential approaches to their management. J Pharmacogn Phytochem 2019;8:621-6.
7. Kausar R, Iram S, Ahmad KS, Jaffri SB. Molecular characterization of Fusarium solani and Fusarium oxysporum phyto-pathogens causing mango maturity malconformation. Arch Phytopathol Plant Prot 2021;54:1372-90. [DOI]
8. Summanwar AS, Rayeehandhuri SP, Pathak SC. Association of fungus Fusarium moniliformae sheld with the malformation in mango (Mangifera indica L.). Indian J Phytopathol 1966;19:227-8.
9. Varma A, Lele VC, Raychauduri SP, Ram A, Sang A. Mango malformation: A fungal disease. J Phytopathol 1974;70:254-7. [DOI]
10. Nelson PE, Tousson TA, Marasas WFO. Fusarium Species: An illustrated manual for identification. Pennsylvania: The Pennsylvania State University Press; 1983. p. 123.
11. Ansari MW, Bains G, Shukla A, Pant RC, Tuteja N. Low temperature stress ethylene and not Fusarium, might be responsible for mango malformation. Plant Physiol Biochem 2013;69:34-8. [DOI]
12. Marasas WFO, Ploetz RC, Wingfield MJ, Wingfield BD, Steenkamp ET. Mango malformation disease and the associated Fusarium species. Phytopathology 2006;96:667-72. [DOI]
13. Freeman S, Shtienberg D, Maymon M, Levin AG, Ploetz RC. New insights into mango malformation disease epidemiology lead to a new integrated management strategy for subtropical environments. Plant Dis 2014;98:1456-66. [DOI]
14. Britz H, Steenkamp ET, Coutinho TA, Wingfield BD, Marasas WFO, Wingfield MJ. Two new species of Fusarium section Liseola associated with mango malformation. Mycologia 2002;94:722-30. [DOI]
15. Wu J, Liu F, Zhan R, Li G, Zhao Y, Jinmei C, He Y. Development of a sensitive molecular detection assay for mango malformation disease caused by Fusarium mangiferae. Biotechnol Lett 2016;38:835-40. [DOI]
16. Cohen Y, Belausov E, Maymon M, Elazar M, Shulman I, Saada D, Shtienberg D, Freeman S. Fusarium mangiferae localization in planta during initiation and development of mango malformation disease. Plant Pathol 2017;66: 924-33. [DOI]
17. Vrabka J, Niehaus EM, Münsterkötter M, Proctor RH, Brown DW, Novák O, Pěnčik A, Tarkowská D, Hromadová K, Hradilová M, et al. Production and role of hormones during interaction of Fusarium species with maize (Zea mays L.) seedlings. Front Plant Sci 2019; 9:1936. [DOI]
18. Usha K, Singh B, Kamil D. Hormonal profiling of the Fusarium mangiferae infected mango buds in relation to mango malformation. Sci Hortic 2019;254:148-54. [DOI]
19. Nirenberg HI., O’Donnell K. New Fusarium species and combinations within the Gibberella fujikuroi species complex. Mycologia 1998;90:434-58. [DOI]
20. Gardes M, Bruns TD. ITS primers with enhanced specificity for basidiomycetes‐application to the identification of mycorrhizae and rusts. Mol Ecol 1993;2:113 -8. [DOI]
21. White TJ, Bruns T, Lee S, Taylor JW. Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics. In: Innis MA, Gelfand DH, Sninsky JJ, White TJ, editors. PCR protocols: a guide to methods and applications. New York: Academic Press, Inc.; 1990. p. 315-322. [DOI]
22. O’Donnell K, Cigelnik E. Two divergent intragenomic rDNA ITS2 types within a monophyletic lineage of the fungus Fusarium are nonorthologous. Mol Phylogenet Evol 1997;7:103-16. [DOI]
23. Glass NL, Donaldson GC. Development of primer sets designed for use with the PCR to amplify conserved genes from filamentous ascomycetes. Appl Environ Microbiol 1995;61:1323-30. [DOI]
24. Mirghasempour SA, Studholme DJ, Chen W, Cui D, Mao B. Identification and characterization of Fusarium nirenbergiae associated with saffron corm rot disease. Plant Dis 2022;106:486-95. [DOI]
25. Kumar S, Stecher G, Tamura K. MEGA7: Molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol Biol Evol 2016;33:1870-4. [DOI]
26. Kimura M. A simple method for estimating evolutionary rates of base substitutions through comparative studies of nucleotide sequences. J Mol Evol 1980;16:111-20. [DOI]
27. Wei G, Zhao W, Hu A, Ren M, Huang Y, Xu H. Identification of a new Pathogenic fungi causing sorghum leaf spot disease and its management using natural product and microorganisms. Microorganisms 2023;11:1-13. [DOI]
28. Tahir M., Iram S, Ahmad KS, Jaffri SB. Developmental abnormality caused by Fusarium mangiferae in mango fruit explored via molecular characterization. Biologia 2020;75: 465-73. [DOI]
29. Liu F, Wu JB, Zhan RL, Ou XC. Transcription profiling analysis of mango-Fusarium mangiferae interaction. Front Microbiol 2016;7:1-11. [DOI]