Jun Hyuk Park1 , Young-Joon Choi12* , and Hyeon-Dong Shin3
1Department of Biological Science, Kunsan National University, Gunsan 54150, Korea
2Center for Convergent Agrobioengineering, Kunsan National University, Gunsan 54150, Korea
3Division of Environmental Science and Ecological Engineering, Korea University, Seoul 02841, Korea
*Correspondence to yjchoi@kunsan.ac.kr
Korean Journal of Mycology (Kor J Mycol) 2024 December, Volume 52, Issue 4, pages 325-329.
https://doi.org/10.4489/kjm.520411
Received on December 09, 2024, Revised on December 12, 2024, Accepted on December 12, 2024, Published on Dec 30, 2024.
Copyright © The Korean Society of Mycology.
This is an Open Access article which is freely available under the Creative Commons Attribution-NonCommercial 4.0 International License (CC BY-NC) (https://creativecommons.org/licenses/by-nc/4.0/).
Since the 1980s, an invasive plant, Symphyotrichum subulatum, has become widely naturalized in Korea, competing with native plants and disrupting ecosystems. Since powdery mildew symptoms were initially observed on S. subulatum on a riverside in Busan in 2007, infected plants have been continuously observed. Morphological examination and phylogenetic analysis confirmed Golovinomyces ambrosiae as the causal agent. This study reports the first occurrence of powdery mildew caused by G. ambrosiae on S. subulatum in Korea. Given that this pathogen completes its life cycle by developing its sexual stage in Korea, it poses a potential threat to S. subulatum and other Asteraceae plants.
Asteraceae, Golovinomyces cichoracearum, Phylogenetic analysis, Symphyotrichum subulatum
The invasive wild plant Symphyotrichum subulatum (Michx.) G.L. Nesom is native to the tropical regions of America. This plant was accidentally introduced to Korea in the 1980s and has since become widely naturalized, outcompeting indigenous plants and disrupting native ecosystems [1]. In November 2007, S. subulatum plants showing symptoms of powdery mildew were first observed at a riverside in Busan (35°12’47″N 128°59’29″E), Korea. Between 2007 and 2020, four powdery mildew-infected specimens of S. subulatum were collected and deposited in the Korea University Herbarium (KUS-F). Initially, the infection appeared as circular to irregular white patches on the upper surfaces of the leaves, which gradually spread to form a powdery growth on both sides of the leaves, as well as on the stems and inflorescences (Fig. 1A and 1B). The affected leaves exhibited a purple-to-brown discoloration.
For morphological analysis, fungal structures from infected leaves were mounted in distilled water and observed under a bright-field microscope (Olympus BX50, Olympus, Tokyo, Japan). Conidiophores, conidia, and appressoria were measured, and at least 20 measurements were taken for each structure. Morphological examination revealed that the appressoria on the mycelia were nipple-shaped. The conidiophores were simple at 97.4 to 164.3 (av. 130.5) × 9 to 13 (av. 10.5) μm and produced 2 to 4 immature conidia in chains with a sinuate outline (Fig. 1D and 1E). Foot cells of conidiophores were straight and 41.4 to 75.9 (av. 58.4) μm long (Fig. 1C to 1E). Conidia were hyaline, ellipsoid to barrel-shaped, 29.5 to 39 (av. 33.6) × 13.3 to 18 (av. 15.7) μm (a length/width ratio = 1.5 to 2.3), lacked distinct fibrosin bodies, and produced a germ tube at the subterminal position, with reticulate wrinkling of the outer walls (Fig. 1F to 1H). Chasmothecia were amphigenous and caulicolous, scattered to gregarious, subglobose, and 97.6 to 132 (av. 111.5) μm in diameter. Peridium cells were irregularly shaped, ranging from polygonal to daedaleoid, and 8.7 to 16 (av. 12.5) μm in diameter.

Fig. 1. Powdery mildew symptoms (A and B) and morphological features (C–J) of Golovinomyces ambrosiae affecting Symphyotrichum subulatum. (A) Powdery mildew symptoms on the inflorescence and young stems of S. subulatum, showing white fungal growth. (B) Symptoms on leaves characterized by white, powdery patches. (C–E) Conidiophore with foot cell and immature conidia in chains. (F) Conidia with ellipsoid to barrel shape, with a germ tube formed at the subterminal position (G) and with reticulate wrinkling on the outer wall (H). (I) Chasmothecium with appendages and developing ascospores, indicating sexual reproductive structures. (J) Close-up view of developing asci with ascospores inside a chasmothecium.
Appendages were numerous, arising from the lower half of the chasmothecium, mycelioid, and 0.5–2 times longer than the chasmothecial diameter, with a width of 4–8 (av. 5.9) μm. Initially, the appendages were hyaline but later became yellowish to medium brown (Fig. 1I). The asci were usually numbered 6–11, obovoid to clavate saccate, and 62 to 87 (av. 72.2) × 44 to 66 (av. 55.4) μm, typically containing two ascospores (Fig. 1I and 1J). The ascospores were colorless, ellipsoid to ovoid, and 13 to 22.3 (av. 17) × 10 to 18 (av. 13.5) μm (Fig. 1J). These morphological features are consistent with those described for Golovinomyces ambrosiae (Schwein.) U. Braun & R. T. A. Cook [2,3].
For molecular phylogenetic analysis, genomic DNA was extracted from mycelia and conidia scraped from infected leaves using a MagListo 5M Plant Genomic DNA Extraction Kit (Bioneer, Daejeon, Korea). The internal transcribed spacer (ITS1-5.8S-ITS2), the 5′-end of the 28S intergenic spacer (IGS) regions of rDNA, and glyceraldehyde-3-phosphate dehydrogenase (GAPDH) gene were amplified by polymerase chain reaction (PCR) using the primer sets PM10/ITS4, PM3/TW14, IGS-12a/NS1R, and GoGPD-F/ GoGPD-R, respectively [4–7]. PCR products were purified using an AccuPrep PCR Purification Kit (Bioneer) and sequenced by Macrogen, Inc. (Seoul, Korea). The obtained sequences were edited using DNAStar software (Madison, Wis., USA) version 5.05 and deposited in GenBank (Table 1).
Table 1. Powdery mildew specimens of Symphyotrichum subulatum in Korea
| Herbarium no. | Date | Geographic location | GenBank accession numbers | |||
|---|---|---|---|---|---|---|
| ITS | LSU | IGS | GAPDH | |||
| KUS-F27140 | Oct 2012 | Seoul | PQ680183 | – | PQ683339 | – |
| KUS-F27257 | Nov 2012 | Jeju | PQ680182 | PQ680179 | PQ683336 | PQ683341 |
| KUS-F29331 | Jul 2016 | Jindo | PQ680181 | – | PQ683337 | – |
| KUS-F31878 | Jul 2020 | Seoul | PQ680184 | PQ680180 | PQ683338 | PQ683340 |
ITS: internal transcribed spacer; LSU: large-subunit rDNA; IGS: intergenic spacer; GAPDH: glyceraldehyde-3phosphate dehydrogenase.
A Basic Local Alignment Search Tool (BLAST) search revealed that the ITS, 28S, IGS rDNA, and GAPDH sequences shared 100% similarity with reference sequences of G. ambrosiae (MG704839 for ITS, AB769427 for the large-subunit rDNA (LSU), MZ614729 for IGS, and ON360701 for GAPDH). Phylogenetic analysis was performed using sequences from the ITS, 28S, IGS regions, and GAPDH gene. These regions provide comprehensive information on genetic variability, allowing accurate identification and differentiation of closely related fungal species. The sequences of each marker were aligned using MAFFT version 7 [8], and a phylogenetic tree was constructed using the maximum likelihood (ML) method in MEGA 11 [9]. Bootstrap analysis was performed with 1,000 replicates to evaluate the robustness of the inferred phylogenetic relationships. Based on the morphological characteristics and molecular analyses, the causal agent of powdery mildew on S. subulatum was identified as G. ambrosiae (Fig. 2).
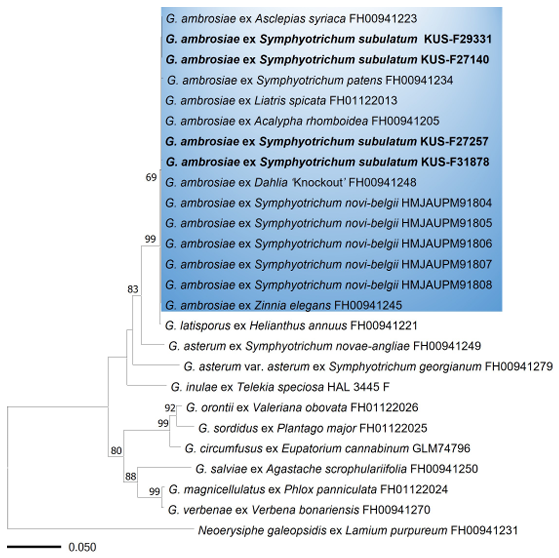
Fig. 2. Maximum likelihood tree of Golovinomyces species inferred from concatenated ITS, LSU, IGS rDNA, and GAPDH sequences. Bootstrap values higher than 60% are indicated above the branches. The blue box represents Golovinomyces ambrosiae. All Korean specimens sequenced in this study are shown in bold. ITS: internal transcribed spacer; LSU: large-subunit rDNA; IGS: intergenic spacer; GAPDH: glyceraldehyde-3-phosphate dehydrogenase.
The powdery mildew of S. subulatum (previously known as Aster subulatus), or its related species, S. squamatum, associated with Golovinomyces cichoracearum s. lat. were reported in Australia, New Zealand, France, Iraq, Israel, and Japan [10]. Previous phylogenetic studies have revealed that G. ambrosiae on S. novi-belgii [9] and S. patens [11] clustered within the same clade but were distinct from G. asterum on S. novae-angliae and G. asterum var. asterum on S. georgianum [11], thus confirming that Symphyotrichum spp. are affected by G. ambrosiae and G. asterum. This study verified that S. subulatum in Korea was specifically infected with G. ambrosiae. To our knowledge, this is the first report of powdery mildew disease caused by G. ambrosiae on S. subulatum in Korea. In addition, we discovered the sexual stage of G. ambrosiae, indicating that the pathogen can complete its life cycle and reproduce sexually in Korea, posing a potential threat to S. subulatum and other Asteraceae plants.
The authors declare that they have no potential conflicts of interest.
None.
1. Park SH, Lee YM, Jeong SY, Jeong SS, Oh SH, Yang JC, Jang GS, Hwang HS, Lee HJ, Jang J. Fieldguide naturalized plants of Korea. Pocheon: Korea National Arboretum; 2012.
2. Braun U, Cook RTA. Taxonomic manual of the Erysiphales (powdery mildews). Utrecht: Centraalbureau voor Schimmelcultures; 2012.
3. Qiu PL, Liu SY, Bradshaw M, Rooney-Latham S, Takamatsu S, Bulgakov TS, Tang SR, Feng J, Jin DN, Aroge T, et al. Multi-locus phylogeny and taxonomy of an unresolved, heterogeneous species complex within the genus Golovinomyces (Ascomycota, Erysiphales), including G. ambrosiae, G. circumfusus and G. spadiceus. BMC Microbiology 2020;20:51. [DOI]
4. White TJ, Bruns T, Lee S, Taylor J. Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics. In: Innis IMA, Gelfand DH, Sninsky JJ, White TJ, editors. PCR protocols: a guide to methods and applications. San Diego: Academic Press; 1990. p. 315-22. [DOI]
5. Carbone I, Kohn LM. A method for designing primer sets for speciation studies in filamentous Ascomycetes. Mycologia 1999;91:553-6. [DOI]
6. Bradshaw M, Tobin PC. Sequencing herbarium specimens of a common detrimental plant disease (powdery mildew). Phytopathology 2020;110:1248-54. [DOI]
7. Park JH, Choi YJ. Designing a novel primer set for GAPDH gene to enhance taxonomic and phylogenetic studies of Golovinomyces species. Mycobiology 2024;52:231-5. [DOI]
8. Katoh K, Standley DM. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol Evol 2013;30:772-80. [DOI]
9. Tamura K, Stecher G, Kumar S. MEGA11: molecular evolutionary genetics analysis version 11. Mol Biol Evol 2021;38:3022-7. [DOI]
10. Farr DF, Rossman AY. Fungal Databases, Syst. Mycol [Internet]. Washington DC: US Department of Agriculture, Agricultural Research Service; 2024 [cited 2024 Dec 5]. Available from https://nt.ars-grin.gov/fungaldatabases/.
11. Bradshaw MJ, Braun U, Pfister DH. Phylogeny and taxonomy of the genera of Erysiphaceae, part 1: Golovinomyces. Mycologia 2022;114:964-93. [DOI]