Do Yoon Lim1, Han Na Park2, Jea Hyuk Yoo2, Siyeon Lim1, Bomi Lee1, Jiyoung Min1, Sun Ha Kim1, Inha Lee2, and Sang-Keun Oh1*
1Department of Applied Biology, College of Agriculture & Life Sciences, Chungnam National University, Daejeon 34134, Korea
2Chungcheong-nam do Agricultural Research & Extension, Nonsan, 32418, Korea
*Correspondence to sangkeun@cnu.ac.kr
Korean Journal of Mycology (Kor J Mycol) 2024 December, Volume 52, Issue 4, pages 341-348.
https://doi.org/10.4489/kjm.520413
Received on December 05, 2024, Revised on December 13, 2024, Accepted on December 13, 2024, Published on Dec 30, 2024.
Copyright © The Korean Society of Mycology.
This is an Open Access article which is freely available under the Creative Commons Attribution-NonCommercial 4.0 International License (CC BY-NC) (https://creativecommons.org/licenses/by-nc/4.0/).
Strawberry Fusarium wilt, caused by Fusarium oxysporum f. sp. fragariae (FOF), poses a significant threat to strawberry production. FOF secretes effector proteins called “secreted in xylem” (SIX) into the plant’s xylem, disrupting the defense responses in the plant. In this study the proteins SIX6a and SIX6b in FOF races 1 and 2, showing race-specific expression patterns, were identified. These genes encode effector proteins with a conserved signal peptide that is typical of fungal effectors. Differential expression in susceptible strawberries suggests that FOF races regulate these effectors differently, thereby contributing to variations in pathogenicity. Understanding these effectors is crucial for the development of racespecific resistance strategies to manage Fusarium wilt in strawberries.
Fungal effectors, Fusarium oxysporum f. sp. fragariae, Fusarium wilt, SIX gene, Strawberry
Strawberry (Fragaria × ananassa), an octoploid plant, is one of the most widely cultivated fruits worldwide. However, strawberry plants are susceptible to various pathogens, which leads to significant production losses [1]. One such pathogen is Fusarium oxysporum (FO), which causes Fusarium wilt. This pathogen has a broad host range among important crops, but different strains are highly host specific and are grouped into formae speciales (f. sp.) [2]. The strain responsible for Fusarium wilt in strawberries is Fusarium oxysporum f. sp. fragariae (FOF) [3]. FOF invades the roots, causes discoloration, multiplies in the xylem, disrupts nutrient movement, and ultimately kills the plant [4,5]. The pathogen produces chlamydospores that survive in unfavorable conditions, spread through the stolons, and infect firstgeneration daughter plants, making Fusarium wilt the most serious strawberry disease [5,6].
During infection, F. oxysporum secretes effectors into the host cells, thus facilitating colonization by modifying the immune response [7]. A group of small, cysteine-rich secreted proteins, known as “secreted in xylem” (SIX), have been identified in infected plants [8–10]. These proteins, present in various Fusarium species, serve as markers to distinguish different formae speciales (f. sp.) and races based on their presence or sequence variations. For example, F. oxysporum f. sp. lycopersici (FOL) race 1 lacks the SIX4 gene, and differences in SIX3 distinguish races 2 and 3 [11]. In F. oxysporum f. sp. cubense (FOC), the SIX8 gene is used for race identification. Research on Fusarium species is ongoing, with the aim of understanding their genetic diversity, pathogenicity, and fungicide response, with the ultimate goal of breeding plants resistant to Fusarium diseases [5,7,12–15].
Recent studies on FOF have identified two distinct symptoms, suggesting the presence of wilt- and yellows-causing isolates [10]. SIX6 is found in yellows-causing isolates, whereas the wilt-causing isolates contain SIX1a, SIX1b, SIX1c, and SIX13 [7,16–18]. The SIX6 gene is present in FOF race 1 but absent in race 2 isolates [19]. Accordingly, we investigated the presence of the SIX6 gene in five Korean FOF isolates, Fo160609, Fo080701 (race 1), Fo160403, Fo160618, and SK1 (race 2), which were obtained from the Agricultural Research & Extension Center in Nonsan (Table 1). Genomic DNA was extracted from five isolates, and sequences were analyzed to determine the presence of SIX6.
Table 1. Fusarium oxysporum f. sp. fragariae isolates obtained from strawberry fields in this study
| Isolate | Cultivar | Source | Year | Location |
|---|---|---|---|---|
| Fo160403 | Sulhyang | Crown | 2016 | Nonsan, CN |
| SK1 | Sulhyang | Crown | 2016 | Hongseong, CN |
| Fo160618 | Sulhyang | Crown | 2016 | Hongseong, CN |
| Fo080701 | Redpearl | Crown | 2008 | Ganghwa, GG |
| Fo160609 | Sulhyang | Crown | 2016 | Nonsan, CN |
CN: Chungcheongnam-do (South Chungcheong Province, Korea); GG: Gyeonggi-do (Gyeonggi Province, Korea).
Five FOF isolates were cultured [13], and mycelia were harvested for genomic DNA extraction. Mycelia were filtered through miracloth and collected in a 50-mL Falcon tube using a sterilized spatula. The samples were either used immediately for DNA extraction or frozen in liquid nitrogen and stored at −80℃. Genomic DNA was extracted with minor modifications to the protocol [20]. Mycelia were ground in liquid nitrogen, resuspended in extraction buffer (50 mM Tris-HCl, 50 mM EDTA, 3% SDS, and 1% 2-mercaptoethanol), and treated with RNase. After vortexing and incubating at 65℃ for 1 h, chloroform was added, followed by centrifugation. The upper aqueous phase was transferred and a 24:1 mixture of chloroform and isoamyl alcohol was added, followed by centrifugation. The DNA was precipitated with 3 M NaOAc and isopropanol, washed with ethanol, and dried. The DNA pellet was resuspended in Tris-EDTA (SmartGene, Daejeon, Korea) buffer and stored at −20℃.
The pathogenicity of the FOF race 1 and race 2 isolates was assessed by inoculating susceptible strawberry plants with spore suspensions (1 × 106 spores/mL) and planting them in autoclaved sand in a growth chamber. Roots from five plants were harvested at 0, 2, 4, 6, 8, and 10 days after inoculation (dai) and flash-frozen in liquid nitrogen. RNA was extracted using Trizol® reagent, and first-strand cDNA was synthesized. Real-time reverse-transcription polymerase chain reaction (RT-PCR) was performed to quantify SIX gene expression, which was analyzed on a Roche LightCycler 480 using SYBR Green I Master Mix (Bio-Rad CFX96TM Real-time System platform, Berkeley, USA). The SIX6 gene was originally identified in strawberry (Fragaria × ananassa) plants following FOF infection [17,18]. SIX6 acts as an avirulence factor (AvrFW1) for FOF race 1 by specifically interacting with a resistance gene located at the FW1 locus in strawberry cultivars [19]. Using the AvrFW1 sequence provided by Dillar-Ermita et al. [19], we successfully cloned SIX6. We then analyzed nine candidate SIX6 genes from five FOF race 1 and race 2 isolates from Korea using FOF-specific primers (FOFra) and primers specific for SIX6a and SIX6b. The results showed the presence of 600- and 700-bp bands for races 1 and 2, respectively (Fig. 1A).
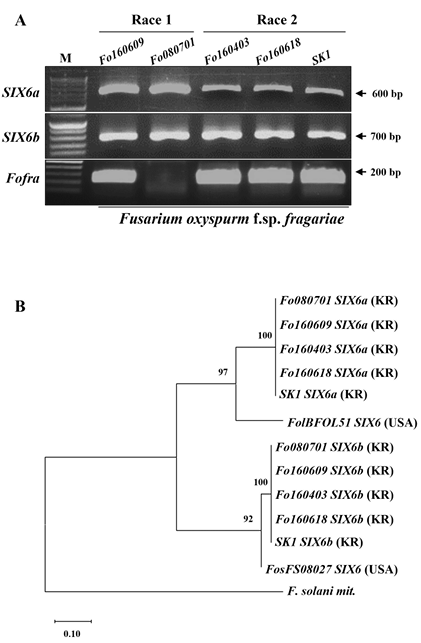
Fig. 1. Identification of the SIX6a and SIX6b gene from 5 different Fusarium oxysporum f. sp. fragariae (FOF) race 1 and race 2 isolates [13]. (A) Multiplex polymerase chain reaction (PCR) with FOFspecific primers(FOFra), a specific primer of SIX6a and SIX6b gene. The 600 bp and 700 bp bands were detected in FOF race 1 and race 2. (B) Maximum likelihood tree for 5 FOF isolates from diseased Korea strawberry, based on an alignment of SIX6a and SIX6b sequences. Numbers represent bootstrap percentage values from 1,000 replicates. Scale bar indicates 0.10 substitutions per site. The tree is rooted through three F. solani isolates (outgroup).
The SIX6 gene has been identified in race 1 yellows-fragariae isolates [19]. To determine whether SIX6 was present in Korean isolates [13], we designed primers for SIX6 and performed PCR on two race 1 isolates and three race 2 isolates. We identified two homologous genes, SIX6a and SIX6b, in race 1 and race 2 isolates (Fig. 1A). Both genes exhibited 100% identical nucleotide sequences with no variations. Basic Local Alignment Search Tool (BLAST) analysis also revealed that SIX6 gene sequences in Korean isolates matched those of yellows-fragariae isolates from other countries. The sequencing results confirmed that SIX6 contains a conserved signal peptide at the N-terminus.
The phylogenetic tree was constructed using MEGA11 with a bootstrap value of 1,000, employing the maximum likelihood method and the Kimura 2-parameter model [21]. The tree was based on the nucleotide sequences of SIX6a and SIX6b from five Korean isolates and two American isolates. The FOF SIX6a and SIX6b genes exhibited 52.8% nucleotide sequence identity. SIX6a homologs from the f ive Korean FOF isolates demonstrated 75.0% sequence identity with the closest homolog derived from the FOL isolate BFOL-51. In contrast, SIX6b homologs from the same five Korean isolates [13] showed significantly higher sequence identity (98.1%) with the closest homolog found in F. oxysporum f. sp. sesami isolate FS08027 (Fig 1B).
To determine whether SIX6a and SIX6b are also expressed during the infection stage, we inoculated in FOF race 1 and race 2 isolates that inoculated into the cultivar ‘Sulhyang’ at a concentration of 1 × 106 spores/mL, and the disease severity was investigated [13]. Both the strains showed yellowing and wilting symptoms in the infected plants. Race 2 exhibited more severe wilting symptoms than did race 1 (Fig. 2A). Second, RNA was collected from FOF race 1-and race 2-infected strawberry plants at 2, 4, 6, 8, and 10 dai. The expression of SIX6a and SIX6b was monitored using RT-PCR, and SIX6a and SIX6b transcripts were detected at selected time points in the infected plants. Interesting results were obtained from the inoculation experiments with FOF race 1 and race 2 isolates. In the FOF race 1 isolate, the expression of both SIX6a and SIX6b began to increase at 2 dai, with the highest expression levels observed at 6 dai. Furthermore, there were no significant differences in the expression levels of these two genes. In contrast, the FOF race 2 isolate exhibited distinct expression patterns: the expression of SIX6a increased on day 2 but then declined, and SIX6b showed no detectable expression initially but exhibited high expression levels on day 10 (Fig. 2B and 2C).
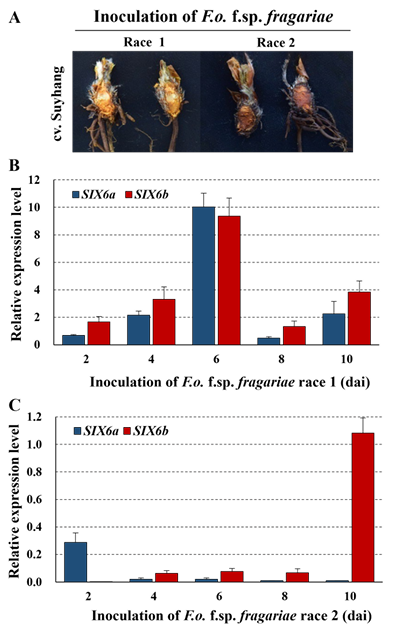
Fig. 2. Pathogenicity of Fusarium oxysporum f. sp. fragariae (FOF) race 1 and race 2 isolates [13] on cultivar Sulhang of strawberry using the root dip inoculation method. Photograph of symptoms were taken 30 days after inoculation (dai) (A). Expression of Secreted In Xylem (SIX6a and SIX6b) genes in strawberry roots infected with FOF race 1 (B) and race 2 (C) isolates as determined by reverse transcription qPCR of RNA. Expression was calculated relative to the β-Tubulin and the mean value displayed [Error bars represent standard error of the mean (SEM)].
The sequences of the two genes in Korean isolates showed no differences [13]. We hypothesized that the expression patterns of these genes would differ between races 1 and 2 when FOF was inoculated onto strawberry roots. Heavy colonization by this pathogen was observed in the vascular tissue of the susceptible cultivar ʻCarmarosa’ during the early stages of infection [20]. After FOF inoculation, samples were collected for up to 10 dai. Reverse-transcription quantitative polymerase chain reaction (RT-qPCR) results revealed that both SIX6a and SIX6b were highly expressed when race 1 was inoculated onto the susceptible cultivar. In contrast, neither of the two genes was expressed when race 2 was inoculated.
SIX1a, SIX1b, SIX1c, SIX6, and SIX13 have been identified in FOF [7,16,17,18]. However, five Korean FOF isolates contained two homologs of SIX6a and SIX6b, whereas FOF isolates from California possessed only one SIX6 gene [7,17,18]. Notably, we did not detect the presence of SIX1a, SIX1b, SIX1c, or SIX13 in the Korean isolates, in contrast to FOF isolates from Australia and Spain, which harbored these genes [16,18]. Furthermore, race 1 isolates of FOF from California exclusively contained SIX6, whereas race 2 isolates did not [19]. In contrast, the race 1 and race 2 FOF isolates from Korea possessed two SIX6 genes. This finding suggests that FOF strains exhibit different combinations of SIX genes, depending on geographic location. However, no sequence differences were observed between Korean and California isolates. The expression patterns of SIX genes during pathogen inoculation of susceptible strawberry roots were also similar between the Korean and California race 1 isolates [7,17,18]. Specifically, in race 1, both SIX6a and SIX6b were highly expressed during the early stages of inoculation. These results align with the observation that the vascular tissue of the root in the susceptible cultivar ʻCarmarosa’ is heavily colonized by the pathogen in the early stages of infection [20]. This suggests that FOF race 1 secretes SIX6 to facilitate strawberry root vascular tissue colonization.
In the Korean isolates, race 2 also possessed two SIX6 genes; however, their expression was minimal upon inoculation with susceptible strawberry roots [13]. We hypothesized that race 2 secretes other candidate effector genes responsible for the infection process in strawberry roots. Unlike the Californian isolates, where races 1 and 2 are distinguishable based on their SIX6 gene presence, in the Korean isolates races 1 and 2 can be distinguished by assessing the expression of SIX6a and SIX6b in response to inoculation. We defined race 1 as expressing SIX6a and SIX6b for root colonization, whereas race 2 did not express these genes. This is the first report of two SIX6 gene homologs in FOF isolates from Korea. These f indings provide molecular evidence that this pathogen secretes different virulence factors depending on race when infecting strawberry plants. Additionally, it may be possible to identify race-specific resistance (R) genes in strawberry cultivars that exhibit resistance, and explore the molecular interactions between these R genes and the effector genes of the FOF.
The authors declare that there are no conflicts of interest.
This work was supported by the Rural Development Administration funded by the Korean Government (Project No. RS-2024-00439133), Republic of Korea.
1. Garrido C, Carbu M, Fernandez-Acero FJ, Gonzalez-Rodriguez VE, Cantoral JM. New insights in the study of strawberry fungal pathogens. In: Husaini AM, Mercado JA, editors. Genomics, Transgenics, Molecular Breeding and Biotechnology of Strawberry, Vol. 5. Genes, Genomes and Genomics. Japan: Global Science Books; 2011. p. 24-39.
2. Snyder WC, Hansen HN. The species concept in Fusarium. Am J Bot 1940;27;64-7. [DOI]
3. Winks BL, Williams YN. A wilt of strawberry caused by a new form of Fusarium oxysporum. Queensland J Agric Anim Sci 1965;22:475-9.
4. Gordon TR. Fusarium oxysporum and the Fusarium Wilt Syndrome. Annu Rev Phytopathol 2017;55:23-39. [DOI]
5. Nam MH, Park MS, Kim HG, Yoo SJ. Biological control of strawberry Fusarium wilt caused by Fusarium oxysporum f. sp. fragariae using Bacillus velezensis BS87 and RK1 formulation. J Microbiol Biotechnol 2009;19:520-4. [DOI]
6. Nam MH, Kang YJ, Lee IH, Kim HG, Chun C. Infection of daughter plants by Fusarium oxysporum f. sp. fragariae through runner propagation of strawberry. Korean J Hort Sci Technol 2011;29:273-7.
7. Houterman PM, Cornelissen BJC, Rep M. Suppression of plant resistance gene-based immunity by a fungal effector. PLoS Pathog 2008;4:e1000061. [DOI]
8. Ma LJ, van der Does HC, Borkovich KA, Coleman JJ, Daboussi MJ, Di Pietro A, Dufresne M, Freitag M, Grabherr M, Henrissat B, et al. Comparative genomics reveals mobile pathogenicity chromosomes in Fusarium. Nature 2010;464:367-73. [DOI]
9. Rep M, Meijer M, Houterman PM, van der Does HC, Cornelissen BJC. Fusarium oxysporum evades I-3-mediated resistance without altering the matching avirulence gene. Mol Plant Microbe Interact 2005;18:15-23. [DOI]
10. Rep M, Van Der Does HC, Meijer M, Van Wijk R, Houterman PM, Dekker HL, De Koster CG, Cornelissen BJC. A small, cysteine-rich protein secreted by Fusarium oxysporum during colonization of xylem vessels is required for I-3-mediated resistance in tomato. Mol Microbiol 2004;53:1373-83. [DOI]
11. Lievens B, Houterman PM, Rep M. Effector gene screening allows unambiguous identification of Fusarium oxysporum f. sp. lycopersici races and discrimination from other formae speciales. FEMS Microbiol Lett 2009;300:201-15. [DOI]
12. Jenner BN, Henry PM. Pathotypes of Fusarium oxysporum f. sp. fragariae express discrete repertoires of accessory genes and in-duce distinct host transcriptional responses during root infection. Environ Microbiol 2022;24:4570-86. [DOI]
13. Nam MH, Kim HS, Park MS, Min JY, Kim HT. Genetic diversity, pathogenicity, and fungicide response of Fusarium oxysporum f. sp. fragariae isolated from strawberry plants in Korea. Research in Plant Disease 2020;26:79-87. [DOI]
14. Pincot DDA, Feldmann MJ, Hardigan MA, Vachev MV, Henry PM, Gordon TR, Bjornson M, Rodriguez A, Cobo N, Famula RA, et al. Novel Fusarium wilt resistance genes uncovered in natural and cultivated strawberry populations are found on three non-homoeologous chromosomes. Theor Appl Genet 2022;135:2121-45. [DOI]
15. Pincot DDA, Poorten TJ, Hardigan MA, Harshman JM, Acharya CB, Cole GS, Gordon TR, Stueven M, Edger PP, Knapp SJ. Genome-wide association mapping uncovers Fw1, a dominant gene conferring resistance to Fusarium wilt in strawberry. G3 2018;8:1817-28. [DOI]
16. Henry PM, Pincot DDA, Jenner BN, Borrero C, Aviles M, Nam MH, Epstein L, Knapp SJ, Gordon TR. Horizontal chromosome transfer and independent evolution drive diversification in Fusarium oxysporum f. sp. fragariae. New Phytol 2021;230:327-40. [DOI]
17. Czislowski E, Fraser-Smith S, Zander M, O’Neill WT, Meldrum RA, Tran-Nguyen LTT, Batley J, Aitken EAB. Investigation of the diversity of effector genes in the banana pathogen, Fusarium oxysporum f. sp. cubense, reveals evidence of horizontal gene transfer. Mol Plant Pathol 2018;19:1155-71. [DOI]
18. Jenner BN, Henry PM. Pathotypes of Fusarium oxysporum f. sp. fragariae express discrete repertoires of accessory genes and induce distinct host transcriptional responses during root infection. Environ Microbiol 2022;24:4570-86. [DOI]
19. Dilla-Ermita CJ, Goldman P, Anchieta A, Feldmann MJ, Pincot DDA, Famula RA, Vachev M, Cole GS, Knapp SJ, Klosterman SJ, et al. Secreted in Xylem 6 (SIX6) mediates Fusarium oxysporum f. sp. fragariae race 1 avirulence on FW1-resistant strawberry cultivars. Mol Plant Microbe Interact 2024;37:530-41. [DOI]
20. Lee SB, Milgroom MG, Taylor JW. A rapid, high yield mini-prep method for isolation of total genomic DNA from fungi. Fungal Genetics Reports 1988;35:23-4. [DOI]
21. Tamura K, Stecher G, Kumar S. MEGA11: molecular evolutionary genetics analysis version 11. Mol Biol Evol 2021;38:3022-7. [DOI]