Hyun Uk Cho, Na Young Yoon, HyeongJin Noh, Ye In Kim, and Seong Hwan Kim*
Department of Microbiology, Dankook University, Cheonan 31116, Korea
*Correspondence to piceae@dankook.ac.kr
Korean Journal of Mycology (Kor J Mycol) 2024 December, Volume 52, Issue 4, pages 349-370.
https://doi.org/10.4489/kjm.520414
Received on December 01, 2024, Revised on December 15, 2024, Accepted on December 15, 2024, Published on Dec 30, 2024.
Copyright © The Korean Society of Mycology.
This is an Open Access article which is freely available under the Creative Commons Attribution-NonCommercial 4.0 International License (CC BY-NC) (https://creativecommons.org/licenses/by-nc/4.0/).
Sordariomycetes is the second largest class within the Ascomycota and an ecologically and economically important group in fungal taxonomy. As a result of investigating fungal diversity in the soil of Anseong, Cheonan, Wando, and Jeju Island, we identified six species belonging to Sordariomycetes that had not been previously recorded in Korea. To confirm the taxonomic position of these species, we performed morphological observations and phylogenetic analyses based on the nucleotide sequences of the internal transcribed spacer (ITS), 28S ribosomal RNA gene (LSU rDNA), TEF1-α gene (TEF1-α), β-tubulin gene (β-TUB), and RNA polymerase II gene (RPB2). As a result, six species (Cladorrhinum samala, Fusarium miscanthi, Gymnascella dankaliensis, Microascus gracilis, Papulaspora equi, and Paecilomyces purpureus) were listed as newly confirmed species in Korea.
Soil, Sordariomycetes, Unrecorded fungi
The class Sordariomycetes of the Ascomycota phylum was recently recognized as comprising 45 orders, 167 families, and 1,499 genera [1,2]. This type of fungus exists in various environments, including soil, air, fresh water, and oceans, and in animals and plants [3–7]. The lifestyle of fungi in this class also includes endophytes, parasites, human pathogens, plant pathogens, mushroom pathogens, fish and animal pathogens, and antagonists of bacteria that could be used for biological control agents [7–12]. Additionally, certain groups of Sordariomycetes are useful in industrial application [13].
Geographically, Korea belongs to the mid-latitude temperate climate zone and has four distinct seasons. It is also surrounded by the sea on three sides and consists of inland and islands areas [14]. As Korea has various climates and environments, it can be a valuable place for exploring diverse fungal species. As the fungi of Sordariomycetes are highly diverse in morphology, growth form, and habitat, we are interested in exploring the biodiversity and species distribution of this ecologically and economically important taxon. To study fungal diversity in soil across different geographical regions in Korea, we collected soil samples from the inland regions of Anseong and Cheonan and the island regions of Wando and Jeju. As a result of investigating the fungal diversity of the collected soil, we identified six species of Sordario fungi that had not been recorded in Korea. These species are known as plant pathogens, industrial agents, biological control agents, and metabolite producers. In this study, we report on the molecular and morphological identification and characterization of these species.
Collecting soil samples and isolation of fungi. We collected soil samples in Anseong, Cheonan, Wando and Jeju Island. After removing the external soil to a radius of 10 cm and depth of 10 cm, we used a trowel to collect soil to a depth of 10 cm. Samples soils were sealed in clean sample bags and then used for analysis. We placed 3 g of each of the six soil samples and 30 mL of primary sterile distilled water in a 50-mL tube and mixed them in a vortex mixer for 10 minutes. The mixed solution was diluted up to 100 times using the serial dilution method; moreover, 100 μL of the diluted solution was inoculated onto DG18 (DiCholran glycerol 18% agar, MB cell, Seoul, Korea) and smeared using a glass smear. The plated medium was sealed with parafilm and cultured in an incubator at 25℃ for 7 days. The grown fungi were inoculated onto PDA (Potato Dextrose Agar, Difco, Detroit, MI, USA) using a sterilized inoculation loop and cultured in an incubator at 25℃ for 7 days. Single spore cultures were obtained from PDA-grown fungi and assigned Dankook University Culture Collection (DUCC) numbers. Among the strains assigned DUCC numbers, representative strains were deposited at the National Institute of Biological Resources (NIBR), Incheon, Republic of Korea, where they were given NIBRFGC numbers.
Fungal morphological examination and molecular maker gene analysis. For morphological analysis, the isolated fungi were cultured on PDA, MEA (Malt Extract Agar, MBcell, Seoul, Korea), CYA (Czapek Yeast extract Agar, MBcell, Seoul, Korea), and OA (Oatmeal Agar, Difco, Detroit, MI, USA) media in the dark at 25℃ for 7 days. We observed the morphological characteristics of the pure isolates under a light microscope (BX53: Olympus, Tokyo, Japan) and measured the length and size of the ultrastructure. We extracted genomic DNA from the isolated fungus using a Direct DNA prep kit (NaviBiotech, Cheonan, Korea). The extracted DNA was subjected to PCR (Bio-Rad T100 Thermal Cycler) using the universal primers of internal transcribed spacer (ITS), 28S ribosomal RNA gene (LSU rDNA), TEF1-α gene (TEF1α), β-tubulin gene (β-TUB), and RNA polymerase II gene (RPB2). PCR conditions and primer sequences are provided in Table 1. The amplified PCR products were subjected to 1% agarose gel electrophoresis to confirm DNA amplification and the size of amplified DNA. The PCR amplified product was purified using the High Pure Product Purification Kit (Roche, Indianapolis, IN, USA) and sequenced by Macrogen Corp. (Daejeon, Korea).
Table 1. PCR condition and primer sequences used in this study
| Target region | Primer name | Primer sequence (5’ – 3’) | PCR condition | Reference |
|---|---|---|---|---|
| ITS | ITS1 | GAAGTAAAAGTCGTAACAAGG | 95℃ 5min; 35 cycles: 95℃ 30s, 56℃ 30s, 72℃ 1min; 72℃ 5min | [19] |
| ITS4 | TCCTCCGCTATTGATATGC | [20] | ||
| LSU rDNA | LR0R | ACCCGCTGAACTTAAGC | 95℃ 5min; 35 cycles: 95℃ 30s, 56℃ 30s, 72℃ 1min; 72℃ 5min | [21] |
| LR5 | TCCTGAGGGAAACTTCG | |||
| TEF1-α | TEF1 | GCCATCCTTGGAGATACCAGC | 95℃ 5min; 35 cycles: 95℃ 30s, 55℃ 30s, 72℃ 1min; 72℃ 5min | [22] |
| TEF728 | CATCGAGAAGTTCGAGAAGG | |||
| β-TUB | Bt2a | GGTAACCAAATCGGTGCTGCTTTC | 95℃ 10min; 35 cycles: 95℃ 50s, 52℃ 50s, 72℃ 1min; 72℃ 5min | [23] |
| Bt2b | ACCCTCAGTGTAGTGACCCTTGGC | |||
RPB2 | RPB2-5f | GAYGAYMGWGATCAYTTYGG | 95℃ 10min; 35 cycles: 95℃ 30s; 60℃ 1min; 72℃ 1min; 72℃ 7min | [24] |
| RPB2-7cR | CCCATRGCTTGYTTRCCCAT |
ITS: internal transcribed spacer; LSU rDNA: 28S ribosomal RNA gene; TEF1-α: translation elongation factor 1-α gene; β-TUB: β-tubulin gene; RPB2: RNA polymerase II gene.
Molecular phylogenetic analysis. The determined nucleotide sequences were compared for homology with the sequences of marker genes of fungi in the National Center for Biotechnology Information (NCBI, 2023) database using GenBank BLASTN (http://blast.ncbi.nlm.nih.gov/) tool and the species with the closest nucleotide sequence similarity were searched. To analyze phylogeny of the DUCC strains, multiple sequence alignment was performed using the CLUSTAL W alignment tool in the MEGA 11 program with reference sequences obtained from the GenBank [15]. The reference sequences used in this study are listed in Tables 2–7. We used the aligned sequences to generate a molecular phylogenetic tree of the strains using the Kimura two-parameter model and maximum likelihood (ML) in the MEGA 11 program [16,17]. The reliability of phylogenetic tree branches was analyzed using 1,000 bootstrap analyses [18].
Table 2. Reference sequences used for phylogenetic analysis of strain NIBRFGC000510203
| Scientific name | Strain | Source | Country | ITS | LSU rDNA |
|---|---|---|---|---|---|
| Cladorrhinum brunnescens | CBS 643.75A | – | Netherlands | NR_137152 | NG_073587 |
| Cladorrhinum carnegieae | CBS 150814 | Carnegiea gigantea | USA | NR_197920 | PP791451 |
| Cladorrhinum coprophilum | HGUP191016 | Rosa roxburghii | China | MZ724892 | MZ724986 |
| Cladorrhinum foecundissimum | BCCM 6980 | – | – | KT321080 | KT312993 |
| Cladorrhinum globisporum | LC5415 | Water | China | KU746680 | KU746726 |
| Cladorrhinum hyalocarpum | CBS 322.70 | – | Netherlands | NR_172536 | MH871448 |
| Cladorrhinum intermedium | CBS 433.96 | Soil | India | NR_165594 | MK926859 |
| Cladorrhinum olerum | CBS 120012 | Wombat dung | Australia | – | MT731522 |
| Cladorrhinum queenslandicum | BRIP 63076a | Poa annua | Australia | NR_197571 | PP766573 |
| Cladorrhinum samala | CBS 302.90 | – | – | KT321079 | KT312992 |
| Cladorrhinum samala | INTA-AR 112 | Soybean crop | – | KT321075 | KT312988 |
| Cladorrhinum samala | NIBRFGC000510203 | Soil | Republic of Korea | OQ991278 | OQ991295 |
| Cladorrhinum tomentosum | Francoise Candoussau (S) | Brassica oleracea | Spain | – | KF557691 |
| Podospora bulbillosa | CBS 304.90 | Desert soil | Egypt | NR_077199 | NG_073583 |
| Podospora dacryoidea | INTA-AR 70 | Soybean crop | – | KT321062 | KT312976 |
| Podospora flexuosa | FMR 10415 | Soil | Spain | NR_154757 | NG_058773 |
| Triangularia microsclerotigena | CBS 290.75 | Musa sp. | Turkey | NR_154758 | NG_073584 |
| Triangularia phialophoroides | CBS 301.90 | Desert sand | Egypt | NR_077198 | NG_073586 |
| Valsa nivea | MFLUCC 15-0860 | Salix acutifolia | Russia | KY417737 | KY417771 |
ITS: internal transcribed spacer; LSU rDNA: 28S ribosomal RNA gene.
Strain identified in this study and its accession numbers are indicated in bold.
Table 3. Reference sequences used for phylogenetic analysis of strain NIBRFGC000510209 strain
| Scientific name | Strain | Source | Country | TEF1-α | RPB2 | β-TUB |
|---|---|---|---|---|---|---|
| Fusarium denticulatum | CBS 407.97 | Ipomoea batatas | USA | MN534000 | MN534274 | MN534068 |
| Fusarium pseudocircinatum | LC13677 | Syzygium samarangense | China | MW580543 | MW474489 | MW533822 |
| Fusarium sacchari | CBS 223.76 | Saccharum officinarum | India | MW402115 | KU604309 | MW402313 |
| Fusarium andiyazi | CBS 134430 | – | – | KC954401 | KU604232 | KU603867 |
| Fusarium annulatum | CBS 792.91 | Gladiolus | Netherlands | MW402153 | MW402774 | MW402354 |
| Fusarium brevicatenulatum | CBS 404.97 | Striga asiatica | Madagascar | MN533995 | MN534295 | MN534063 |
| Fusarium concentricum | CBS 102157 | Macaranga pruinosa stem | Malaysia | MW401963 | MW402728 | MW402164 |
| Fusarium ficicrescens | CBS 125178 | Ficus carica | Iran | KU604452 | KT154002 | KP662896 |
| Fusarium foetens | CBS 110286 | Begonia elatior hybrid | Netherlands | MT011001 | MW928825 | MT011049 |
| Fusarium fujikuroi | LC7864 | Poaceae sp. | China | MW580493 | MW474439 | MW533772 |
| Fusarium globosum | CBS 431.97 | Zea mays seed | South Africa | MW402131 | MW402816 | MW402330 |
| Fusarium lactis | NRRL 25200 | – | – | MN193862 | MN193890 | U61551 |
| Fusarium mangiferae | CBS 119853 | Mango | South Africa | MN534016 | MN534270 | MN534140 |
| Fusarium metavorans | LC5933 | Submerged wood | China | MW620178 | MW474703 | MW534057 |
| Fusarium miscanthi | NRRL 26231 | – | – | KU171725 | JX171634 | AF060384 |
| Fusarium miscanthi | LC7503 | Water | China | MW594318 | MW474551 | MW533922 |
| Fusarium miscanthi | NIBRFGC000510209 | Soil | Republic of Korea | PQ611945 | PQ611946 | PQ611947 |
| Fusarium ngaiotongaense | LC13834 | Armeniaca mume | Japan | MW620176 | MW474701 | MW534055 |
| Fusarium nygamai | CBS 749.97 | Sorghum bicolor | Australia | MW402151 | KU604262 | MW402352 |
| Fusarium oxysporum | CBS 221.49 | Camellia sinensis | Southeast Asia | MH484963 | MH484872 | MH485054 |
| Fusarium paranisikadoi | LC2824 | Unidentified grass | China | MW594317 | MW474550 | MW533921 |
| Fusarium proliferatum | CBS 480.96 | Soil | Papua New Guinea | MN534059 | MN534272 | MN534129 |
| Fusarium udum | CBS 747.79 | Cajanus cajan | India | MN534045 | MN534258 | MN534141/td> |
| Fusarium verticillioides | CBS 122159 | – | – | KR071707 | KU604224 | KU603856 |
TEF1-α: translation elongation factor 1-α gene; β-TUB: β-tubulin gene; RPB2: RNA polymerase II gene.
Strain identified in this study and its accession numbers are indicated in bold.
Table 4. Reference sequences used for phylogenetic analysis of strain NIBRFGC000510207
| Scientific name | Strain | Source | Country | ITS | LSU rDNA |
|---|---|---|---|---|---|
| Arthropsis truncata | CBS 584.82 | – | Peru | NR_159641 | NG_056973 |
| Gymnascella afilamentosa | CBS 658.71 | – | USA | NR_160135 | NG_057626 |
| Gymnascella aurantiaca | CBS 405.84 | Mouse dung | Netherlands | OM468610 | OM515116 |
| Gymnascella dankaliensis | NIBRFGC000510207 | Soil | Republic of Korea | OQ991280 | OQ991297 |
| Gymnascella dankaliensis | CBS 352.68 | – | Netherlands | MH859156 | MH870867 |
| Gymnascella dankaliensis | CBS 339.65 | – | India | MH858597 | MH870236 |
| Gymnascella littoralis | CBS 454.73 | – | Canada | NR_155105 | MH872451 |
| Gymnascella nodulosa | CBS 577.63 | – | India | NR_160094 | NG_064039 |
| Gymnascella sp. | BiMM-F285 (SV3) | Dust | Austria | OL527727 | OL527728 |
| Gymnascella stercoraria | LC4076 | Compost | China | NR_160561 | NG 067788 |
| Gymnascella thermotolerans | LC3877 | Soil | China | NR_160560 | NG 067787 |
| Narasimhella hyalinospora | CBS 548.72 | Guinea pig dung | India | NR_130659 | NG_057618 |
| Narasimhella poonensis | CBS 393.71 | – | India | NR_178142 | NG_088077 |
ITS: internal transcribed spacer; LSU rDNA: 28S ribosomal RNA gene.
Strain identified in this study and its accession numbers are indicated in bold.
Table 5. Reference sequences used for phylogenetic analysis of strain NIBRFGC000510198
| Scientific name | Strain | Source | Country | ITS | LSU rDNA | β-TUB |
|---|---|---|---|---|---|---|
| Microascus alveolaris | CBS 139501 | Human | USA | NR_155393 | NG_069390 | KX924263 |
| Microascus alveolaris | CBS 494.70 | Marine sediment | Norway | MH859814 | MH871586 | LN850855 |
| Microascus atrogriseus | CBS 295.52 | Culture contaminant | UK | NR_155415 | KX924030 | KX924265 |
| Microascus campaniformis | CBS 138126 | Human BAL | USA | NR_132937 | HG380495 | LM652606 |
| Microascus cinereus | CBS 365.65 | Soil | India | MH858612 | MH870254 | KX924270 |
| Microascus cirrosus | CBS 217.31 | Prunus sp. | Italy | MH855194 | MH866642 | KX924273 |
| Microascus cirrosus | CBS 267.49 | Sciurus vulgaris | Netherlands | KX923840 | AF400865 | KX924275 |
| Microascus gracilis | CBS 369.70 | Wheat flour | Japan | NR_165206 | HG380467 | KX924297 |
| Microascus gracilis | NIBRFGC000510198 | Soil | Republic of Korea | OP604172 | OP604180 | OP973115 |
| Microascus macrosporus | CBS 662.71 | Soil | USA | LM652423 | LM652517 | LM652636 |
| Microascus pyramidus | CBS 668.71 | Pocket mouse | USA | KX923926 | MH872050 | LN850876 |
| Microascus senegalensis | CBS 594.78 | Skin | Algeria | LN850781 | LN850830 | LN850878 |
| Microascus terreus | CBS 601.67 | Soil | Ukraine | LN850783 | LN850832 | LN850880 |
| Microascus terreus | CBS 665.71 | Soil | USA | KX923938 | – | KX924372 |
| Microascus trigonosporus var. trigonosporus | CBS 150.64 | Allium cepa seed | USA | MH858399 | MH870027 | KX924262 |
| Yunnania carbonaria | MUCL 9027 | Soil | Panama | NR_154546 | HG380462 | LM652695 |
ITS: internal transcribed spacer; LSU rDNA: 28S ribosomal RNA gene; β-TUB: β-tubulin gene.
Strain identified in this study and its accession numbers are indicated in bold.
Table 6. Reference sequences used for phylogenetic analysis of strain NIBRFGC000510208
| Scientific name | Strain | Source | Country | ITS | LSU rDNA |
|---|---|---|---|---|---|
| Apiosordaria hamata | CGMCC 3.15231 | Wetland soil | China | KP878307 | KP878305 |
| Cephalotrichum hinnuleum | CBS 289.66 | Deer dung | Australia | NR_159771 | NG_069723 |
| Microthecium sepedonioides | CBS 265.79 | Soil | – | KP970638 | KP970661 |
| Papulaspora equi | NIBRFGC000510208 | Soil | Republic of Korea | OQ991279 | OQ991296 |
| Papulaspora equi | CBS 128690 | – | USA | MH865116 | MH876556 |
| Papulaspora equi | CBS 573.89 | Ocular lesion | USA | NR_154293 | NG_057961 |
| Papulaspora sp. | Kw207/12 | – | – | HF952115 | HG003579 |
| Paralulworthia halima | CMG 69 | Submerged wood | Portugal | MT235737 | MT235754 |
ITS: internal transcribed spacer; LSU rDNA: 28S ribosomal RNA gene.
Strain identified in this study and its accession numbers are indicated in bold.
Table 7. Reference sequences used for phylogenetic analysis of strain NIBRFGC000512619
| Scientific name | Strain | Source | Country | ITS |
|---|---|---|---|---|
| Aphanocladium album | CBS 401.70 | Myxomycete | Netherlands | AJ292401 |
| Lophiostoma multiseptatum | HHUF 27309 | Dead herbaceous twigs | Japan | NR_138018 |
| Lophiostoma ravennicum | MFLUCC 140005 | Juncus sp. | Italy | NR_148079 |
| Metapochonia rubescens | CBS 352.70 | – | USA | MH859706 |
| Metapochonia rubescens | CBS 464.88 | – | UK | NR_160177 |
| Metarhizium acridum | ARSEF 7486 | – | Niger | NR_132019 |
| Metarhizium album | ARSEF 1942 | – | – | NR_152950 |
| Metarhizium anisopliae | ARSEF 7487 | – | Eritrea | NR_132017 |
| Metarhizium baoshanense | CCTCC M2016589 | Soil | China | NR_166220 |
| Metarhizium brunneum | ARSEF 2107 | Coleoptera | USA | NR_132023 |
| Metarhizium chaiyaphumense | BCC 78198 | – | Thailand | NR_173299 |
| Metarhizium cylindrosporum | ACCC 30114 | – | China | NR_169887 |
| Metarhizium flavoviride | ARSEF 2133 | – | – | NR_131992 |
| Metarhizium frigidum | ARSEF 4124 | – | – | NR_132012 |
| Metarhizium gaoligongense | CCTCC M2016588 | Soil | China | NR_169915 |
| Metarhizium globosum | ARSEF 2596 | – | India | NR_132020 |
| Metarhizium granulomatis | UAMH 11028 | Chameleon liver | Denmark | NR_132013 |
| Metarhizium gryllidicola | BCC 82988 | – | Thailand | NR_175627 |
| Metarhizium guizhouense | CBS 258.90 | – | China | NR_163550 |
| Metarhizium lepidiotae | ARSEF 7488 | – | Australia | NR_132018 |
| Metarhizium majus | ARSEF 1914 | Coleoptera | Philippines | NR_152952 |
| Metarhizium novozealandicum | FI-698 (F10) | Lepidoptera | New Zealand | AF139851 |
| Metarhizium novozealandicum | FI-1125 (DAT-F368) | Soil | Australia | AF139853 |
| Metarhizium phasmatodeae | BCC 49272 | – | Thailand | NR_175628 |
| Metarhizium pinghaense | CBS 257.90 | – | China | NR_077205 |
| Metarhizium robertsii | ARSEF 2575 | – | USA | NR_132011 |
| Metarhizium viride | CBS 348.65 | Chameleo lateralis | Madagascar | NR_111173 |
| Nigelia martialis | RLC707iNat84182994 | Insect-larva | Ecuador | OQ871845 |
| Paecilomyces brunneolus | CBS 370.70 | Cucumis sativus | Japan | NR_149328 |
| Paecilomyces clematidis | BRNU 677844 | Clematis sp. | Czech Republic | NR_182939 |
| Paecilomyces dactylethromorphus | CBS 251.55 | Acetic acid | Brazil | NR_149330 |
| Paecilomyces divaricatus | CBS 284.48 | Mucilage bottle with library paste | USA | NR_145143 |
| Paecilomyces formosus | CBS 990.73B | – | Japan | NR_149329 |
| Paecilomyces fulvus | CBS 146.48 | Bottled fruit | UK | NR_103603 |
| Paecilomyces lagunculariae | CBS 373.70 | Wood | Brazil | NR_145144 |
| Paecilomyces niveus | CBS 100.11 | – | – | NR_144910 |
| Paecilomyces purpureus | GZDXIFR-GZSL-5 | – | – | EF640809 |
| Paecilomyces purpureus | NIBRFGC000512619 | Soil | Republic of Korea | OR742033 |
| Paecilomyces purpureus | DUCC15611 | Soil | Republic of Korea | OR742034 |
| Paecilomyces tenuis | GZUIFR-C43-1 | – | – | NR_184868 |
| Paecilomyces variotii | CBS 101075 | Fruit beverage | Japan | NR_130679 |
| Paecilomyces verticillatus | GZDXIFR-49-01 | – | – | DQ836182 |
| Paecilomyces zollerniae | CBS 374.70 | Wood | Brazil | NR_103602 |
| Papiliomyces shibinensis | GZUH SB13050311 | Lepidopteran pupa | China | NR_154178 |
| Pochonia cordycipiticonsociata | CGMCC:317366 | Larvae | China | KM263567 |
| Pochonia cordycipiticonsociata | P678 | Soil | Poland | OK584612 |
| Pochonia suchlasporia | PP14b | Hevea brasiliensis | Peru | FJ884150 |
| Pochonia suchlasporia | C2S1-2009a | Soil | China | HQ660437 |
| Pochonia suchlasporia | C3S3-2009b | Soil | China | HQ660438 |
| Purpureocillium atypicola | NBRC 9205 | Trapdoor spider | Japan | LC848307 |
| Purpureocillium lavendulum | CBS 128677 | – | Spain | NR_166039 |
| Purpureocillium lilacinum | NRRL 895 | – | – | NR_165946 |
| Purpureocillium sodanum | IBRC-M 30175 | Salt crystals | Iran | KX668542 |
| Purpureocillium takamizusanense | NBRC:110232 | Unidentified cicada | Japan | LC008205 |
| Purpureomyces khaoyaiensis | BCC12687 | Lepidoptera | Thailand | JN049868 |
| Purpureomyces khaoyaiensis | BCC12587 | Homo sapiens | Japan | KX983461 |
| Purpureomyces pyriformis | BCC85348 | Homo sapiens | Japan | MN781927 |
| Purpureomyces pyriformis | BCC85349 | Homo sapiens | Japan | MN781928 |
| Purpureomyces pyriformis | BCC85074 | Homo sapiens | Japan | MN781929 |
ITS: internal transcribed spacer.
Strains identified in this study and their accession numbers are indicated in bold.
Strains examined: Seoun-myeon, Anseong-si, Gyeonggi-do, Republic of Korea (36°55’35.8″N 127° 14’29.4″E), isolated from soil in which pear trees was buried for disease control, collected on January 6, 2023, strain DUCC24252(NIBRFGC000510203).
Macro morphological characteristics (Fig. 1A–D)
The morphology of the colony was observed after 7 d of incubation in the dark condition at 25℃. In all media, the colonies were fully grown to fill the plate. White mycelia initially grow radially in a chrysanthemum-like pattern, eventually turning into a dull light green, with the center remaining white and periphery shifting to a dull light green. Over time, dense cotton-like mycelia appear, and after one month, the color of this cottony mycelium turns buff.
Microscopic morphological characteristics (Fig. 1E–I)
Microsclerotia were attached to thick, dark brown mycelia. They were irregularly shaped, comprising several cells with rounded to polygonal shapes, measuring 79.4–89.4 × 89.8–108.7 μm. Phialides were formed laterally or at the terminal end of the branch, 4.66–8.34 × 2.09–2.85 μm, one conidiogenous opening, bearing conspicuous collarettes. Collarettes size was 1.01–4.3 × 0.6–2.69 μm. Conidia were mostly spherical, often resembling water drops that have a cylindrical with a clump of cut underparts, hyaline, 1.85–2.98 × 2.15–2.85 μm, observed after two months of incubation. Microsclerotia were observed, but teleomorph was not observed.
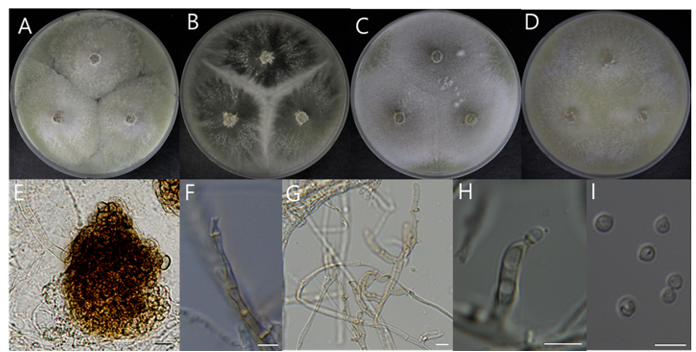
Fig. 1. Morphological characteristics of Cladorrhinum samala NIBRFGC000510203 incubated at 25 ℃ for 7 days. (A) PDA; (B) MEA; (C) CYA; (D) OA; (E) Micro sclerotium; (F) Phialides; (G, H) Conidiophores and phialides; (I) Conidia. Scale bar = 10 μm. PDA: Potato Dextrose Agar; MEA: Malt Extract Agar; CYA: Czapek Yeast extract Agar; OA: Oatmeal Agar.
Molecular characteristics (Fig. 2)
The ITS and LSU rDNA sequences of strain NIBRFGC000510203 were used for phylogenetic analysis. The size of ITS and LSU rDNA sequences were 520 bp and 875 bp, respectively. These two sequences were concatenated and aligned with sequences of other Cladorrhinum species, resulting in a final sequence of 939 bp including gaps. As a result of constructing an ML phylogenetic tree based on the 939 bp concatenated sequence, strain NIBRFGC000510203 formed the same clade with C. samala INTA-AR 112 and C. samala CBS 302.90.
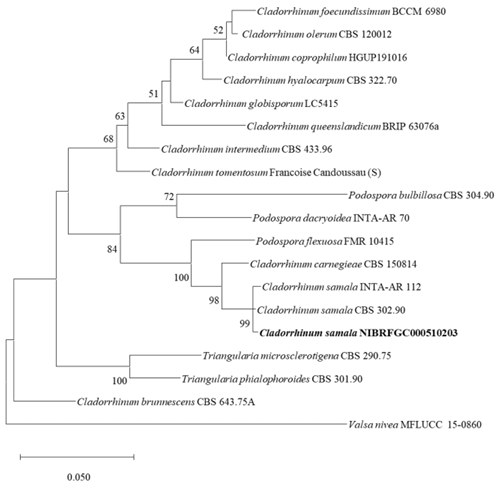
Fig. 2. The phylogenetic tree based on the concatenated nucleotide sequences of the ITS and LSU rDNA using the ML method with the Kimura 2-parameter model. The strain identified in this study is indicated in bold. The number of nodes represents the reliability value through 1,000 bootstrap replicates. Bootstrap values whose node reliability was less than 50 were removed. Valsa nivea MFLUCC 15-0860 was used as an outgroup. ITS: internal transcribed spacer; LSU rDNA: 28S ribosomal RNA gene; ML: maximum likelihood.
Notes
In 1964, Cladorrhinum samala was first reported as an isolate from cow dung in the Chakrata district and characterized by numerous funnel-shaped or cup-shaped collarettes on brown mycelia [25]. Some Cladorrhinum spp. have potential as biological control agents against plant pathogenic fungi and are promising candidates for developing biofertilizers for plant production. Cladorrhinum spp. are also known to produce phytase, an enzyme used in the processing of animal feed [26]. In this study, strain NIBRFGC000510203 was also observed with numerous funnel-shaped or cup-shaped collarlets. Phylogenetic analysis using the combined ITS and LSU rDNA sequences showed that it formed the same clade as C. samala. Based on the results of morphological and molecular analysis, we identified NIBRFGC000510203 as Cladorrhinum samala, a species not previously recorded in Korea.
Strains examined: Seoun-myeon, Anseong-si, Gyeonggi-do, Republic of Korea (36°55’35.8″N 127° 14’29.4″E), isolated from soil in which pear trees was buried for disease control, collected on January 6, 2023, strain DUCC22601(NIBRFGC000510209).
Macro morphological characteristics (Fig. 2A–D)
We observed the morphology of the colony after 7 days of incubation in the dark condition at 25℃. On PDA, 46–47mm diameter radial, circular, slightly fluffy, mycelia tend to white, little purple hyphae in the middle, purple pigment was found. On MEA, 39–40-mm diameter, white mycelia, hyphae at the end were aerial hyphae. On CYA, 45–46-mm diameter, pinkish white, fluffy mycelia. On OA, 49–50mm diameter, white colored, flat, mycelia density lower than other media.
Microscopic morphological characteristics (Fig. 3E–H)
Hyphae were septate, thin walled, mostly cylindrical. Conidiophores were solitary, sometimes lacking a basal septum, 8.5–12.9 × 2.3–2.9 μm, producing one-celled microconidia of two kinds. Pyriform microconidia were one-celled, smooth surface, circular to elliptical with one pointed end like a droplet, 4.38.6 × 2.3–3.3 μm. Fusiform microconidia were one-celled, smooth surface, thin walled, cylindrical shape, 5.5–7.7 × 2.8–6.7 μm. Macroconidia were 2–3 septate, smooth surface, tapering apical cell, 24–26.5 × 3–4.7 μm. Chlamydospores were absent, and teleomorph was not observed.
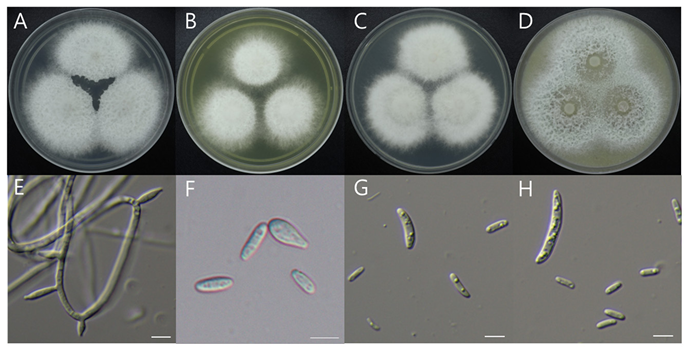
Fig. 3. Morphological characteristics of Fusarium miscanthi NIBRFGC000510209 incubated at 25 ℃ for 7 days. (A) PDA; (B) MEA; (C) CYA; (D) OA; (E) Aerial conidiophores; (F) Pyriform and fusiform microconidia; (G) Fusiform microconidia; (H) Fusiform microconidia and macroconidia. Scale bar = 10 μm. PDA: Potato Dextrose Agar; MEA: Malt Extract Agar; CYA: Czapek Yeast extract Agar; OA: Oatmeal Agar.
Molecular characteristics (Fig. 4)
The sequences of TEF1-α, β-TUB, and RPB2 of strain NIBRFGC000510209 were analyzed. The size of TEF1-α, β-TUB, and RPB2 sequences was 522 bp, 315 bp, and 1,055 bp, respectively; these three sequences were concatenated. The concatenated sequence aligned with sequences of other Fusarium species, resulting in a final sequence of 1,497 bp including gaps. As a result of constructing an ML phylogenetic tree based on the 1,497 bp concatenated sequence, strain NIBRFGC000510209 formed the same clade with F. miscanthi LC7503 and F. miscanthi NRRL 26231.
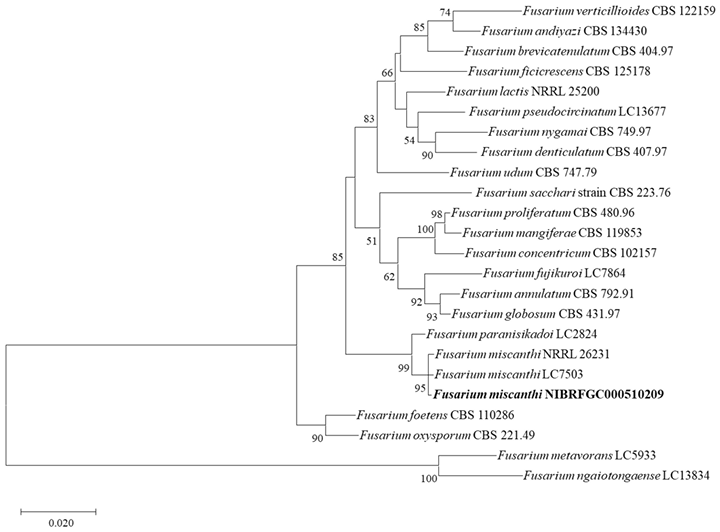
Fig. 4. The phylogenetic tree based on the concatenated nucleotide sequences of the TEF1–α, RPB2, and β-TUB using the ML method with the Kimura 2-parameter model. The strain identified in this study is indicated in bold. The number of nodes represents the reliability value through 1,000 bootstrap replicates. Bootstrap values whose node reliability was less than 50 were removed. Fusarium metavorans LC5933 and Fusarium ngaiotongaense LC13834 were used as outgroups. TEF1-α: translation elongation factor 1-α gene; β-TUB: β-tubulin gene; RPB2: RNA polymerase II gene; ML: maximum likelihood.
Notes
In 1998, Fusarium miscanthi was first reported from straw of Miscanthus sinensis in Denmark and characterized by two forms of microconidia, 3–5 septated macroconidia, and solitary or sporodochial conidiophores. [27]. F. miscanthi causes rhizome rot disease under greenhouse conditions and is also a known cause of ear rot in maize [28,29]. In this study, strain NIBRFGC000510209 also has solitary conidiophore, two forms of microconidia and macroconidia. However, the size of the macroconidia was small, and the number of septa in the macroconidia was 2–3, which was less than in previous reports. Nevertheless, in phylogenetic analysis using the combined TEF1-α, β-TUB and RPB2 sequences, strain NIBRFGC000510209 formed the same clade as F. miscanthi. Based on the results of morphological and molecular analysis, strain NIBRFGC000510209 was Fusarium miscanthi, a species not previously recorded in Korea.
Strains examined: Seonggeo-eup, Seobuk-gu, Cheonan-si, Chungcheongnam-do, Republic of Korea (36°52’23.5″N 127°12’30.2″E), isolated from soil in which pear trees was buried for disease control, collected on July 7, 2022, strain DUCC24354(NIBRFGC000510207).
Macro morphological characteristics (Fig. 5A–D)
We observed the morphology of the colony after 7 d of incubation in the dark condition at 25℃. On PDA, strain NIBRFGC000510207 grew slowly, to 17.62–19.66mm. Conversely, on CYA, it showed the fastest growth at 32.17–32.69 mm. On MEA and OA, colonies were irregular, slightly fluffy, and white to buff colored.
Microscopic morphological characteristics (Fig. 5E–H)
Gymnothecia were yellow to brown colored, spherical, enclosed structure surrounded by a thick wall, 209.57–309.17 × 193.92–269.46 μm. Asci were yellow to orange colored, eight-spored, and 9.05–14.79 × 6.6–13.24 μm. Ascospores were circular or elliptical, one celled, ivory to yellow colored, and 3.16–5.77 × 2.9–5.45 μm. Anamorph was not observed.
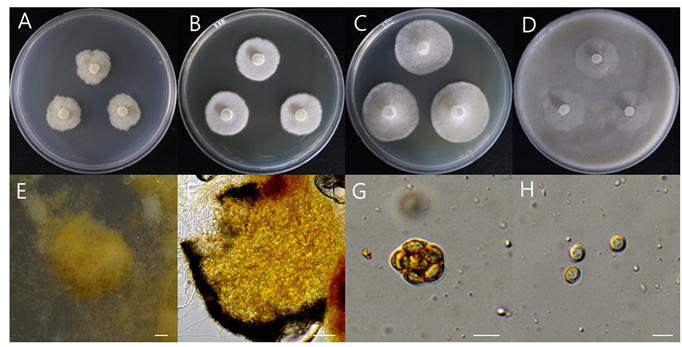
Fig. 5. Morphological characteristics of Gymnascella dankaliensis NIBRFGC000510207 incubated at 25 ℃ for 7 days. (A) PDA; (B) MEA; (C) CYA; (D) OA; (E, F) Gymnothecium; (G) Ascus; (H) Ascospore. Scale bar: E, F = 100 μm, G, H = 10 μm. PDA: Potato Dextrose Agar; MEA: Malt Extract Agar; CYA: Czapek Yeast extract Agar; OA: Oatmeal Agar.
Molecular characteristics (Fig. 6)
The ITS sequences and LSU rDNA sequences of strain NIBRFGC000510207 were used for phylogenetic analysis. The size of ITS and LSU rDNA sequences was 567 bp and 911 bp, respectively. These two sequences were concatenated and aligned with sequences of other Gymnascella species, resulting in a final sequence of 1,138 bp including gaps. As a result of constructing an ML phylogenetic tree based on the 1,138-bp concatenated sequences, strain NIBRFGC000510207 formed the same clade with G. dankaliensis CBS 339.65 and G. dankaliensis CBS 352.68.
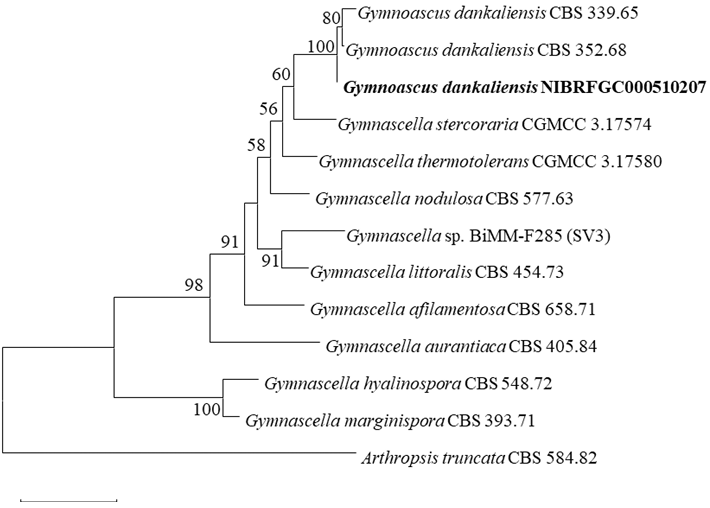
Fig. 6. The phylogenetic tree based on the concatenated nucleotide sequences of the ITS and LSU rDNA using the ML method with the Kimura 2-parameter model. The strain identified in this study is indicated in bold. The number of nodes represents the reliability value through 1,000 bootstrap replicates. Bootstrap values whose node reliability was less than 50 were removed. Arthropsis truncata CBS 584.82 was used as an outgroup. ITS: internal transcribed spacer; LSU rDNA: 28S ribosomal RNA gene; ML: maximum likelihood.
Notes
Gymnascella dankaliensis was first isolated from the hairy skin of a camel in 1937 under the scientific name Trichophyton dankaliense and characterized by round yellow, orange or red brown ascospores encircled by undifferentiated filaments [30]. G. dankaliensis is a keratinophilic fungus that causes superficial infections in humans [31]. Keratinophilic fungi can produce keratinases that degrade keratin materials in the soil [32]. Gymnastatins F and G extracted from the fungus were found to exhibit potent growth inhibition against P388 cancer cell lines [33]. Dankastatin B extracted from the species is involved in the antiproliferative effect on breast cancer cells by targeting C65 in the VDAC3 channel, a mitochondrial poreforming protein [34]. In this study, strain NIBRFGC000510207 represents the morphological characteristics of G. dankaliensis, which exhibits undifferentiated hyphae surrounding yellow ascospores, and formed the same clade with G. dankaliensis CBS 339.65 and G. dankaliensis CBS 352.68 in phylogenetic analysis. Based on the results of morphological and molecular analysis, strain NIBRFGC000510207 was identified as G. dankaliensis-a species not previously recorded in Korea.
Strains examined: Seonghwan-eup, Seobuk-gu, Cheonan-si, Chungcheongnam-do, Republic of Korea 36°55’35.8″N 127°14’29.4″E), isolated from rhizosphere soil of pear tree, collected on November 19, 2022, strain DUCC15381(NIBRFGC000510198).
Macro morphological characteristics (Fig. 6A–D)
The colony morphology was observed after 7 days of incubation in the dark condition at 25℃. Colonies grew slowly, flat, circular colony on all four media (PDA, MEA, CYA, and OA). The colony diameters varied among the four media (9–16 mm). The best media for mycelial growth was CYA. Colony colors were white on PDA, gray in the middle and white outside on MEA, gray in the middle and light brown outside on CYA, and dark gray in the middle and white outside on OA.
Microscopic morphological characteristics (Fig. 7E–S).
Hyphae were hyaline, branched, septate, thin-walled, and 2–3 μm in size. Ascomata are dark, subspherical to spherical, and 160–220 μm in diameter. The development of ascomata took 14 days on OA but more than 3 weeks on the other three media. Asci are oval-shaped, 10.5–16.4 × 6.5–9.1 μm in size, hyaline-filled and transparent, with eight ascospores. Ascospores are 4.3–6.6 × 2.7–3.9 μm in size, oval-shaped, slightly tapered at both ends, pale green, thin-walled. Conidiophores are 6.5–11.6 × 2–3.3 μm in size, cylindrical, internally transparent, pale green, and thin walled. Annellides are 6.4–10.4 × 2.1–3.1 μm in size, pale green, transparent, gourd-shaped, with a transparent interior, and 1–3 at the tip of a conidiophore. Conidia are oval, light green, hyaline and transparent, 3.3–4.6 × 2.6–3.5 μm in size. Microscopic morphological characteristics did not vary substantially among the four media.
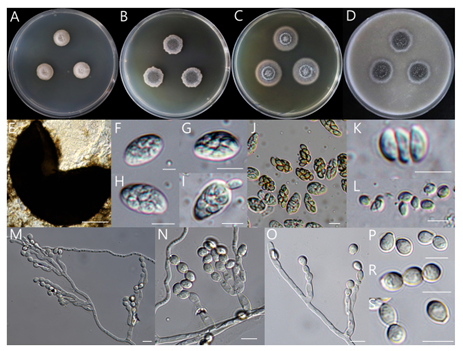
Fig. 7 . Morphological characteristics of Microascus gracilis NIBRFGC000510198 incubated at 25 ℃ for 7 days. (A) PDA; (B) MEA; (C) CYA; (D) OA; (E) Ascocarp; (F–J) Asci; (K, L) Ascospore; (MO) Conidiophore and conidiogenous cells; (P–S) Conidia. Scale bar: E = 100 μm, F–S = 10 μm. PDA: Potato Dextrose Agar; MEA: Malt Extract Agar; CYA: Czapek Yeast extract Agar; OA: Oatmeal Agar.
Molecular characteristics (Fig. 8)
We analyzed the sequences of the ITS, LSU rDNA, and β-TUB. The size of the ITS, LSU rDNA, and β-TUB sequences of strain NIBRFGC000510198 were 485 bp, 909 bp, and 516 bp, respectively. These three sequences were concatenated. The concatenated sequence aligned with sequences of other Microascus species, resulting in a final sequence of 1,332 bp including gaps. As a result of constructing an ML phylogenetic tree based on the concatenated sequences, strain NIBRFGC000510198 formed the same clade with M. gracilis CBS 369.70.

Fig. 8. The phylogenetic tree is based on the concatenated nucleotide sequences of the ITS, LSU rDNA, and β-TUB using the ML method with the Kimura 2-parameter model. The strain identified in this study is indicated in bold. The number of nodes represents the reliability value through 1,000 bootstrap replicates. Bootstrap values whose node reliability was less than 50 were removed. Yunnania carbonaria MUCL 9027 was used as an outgroup. ITS: internal transcribed spacer; LSU rDNA: 28S ribosomal RNA gene; β-TUB: β-tubulin gene; ML: maximum likelihood.
Notes
Microascus gracilis has been isolated mainly from food in Asia and North and South America, as well as from soil in Europe. Morphologically, M. gracilis is characterized by lunate ascospores, conidiophores irregularly branched, and subglobose to ellipsoidal conidia [35]. It caused mycetoma to humans and disseminated infection in a lung transplant patient [36,37]. Strain NIBRFGC000510198 was observed to have morphological characteristics similar to those of M. gracilis, such as lunate ascospores and oval shaped conidia. Based on the results of morphological and molecular analysis, strain NIBRFGC000510198 was identified as Microascus gracilis, a species not previously recorded in Korea.
Strains examined: Yangseong-myeon, Anseong-si, Gyeonggi-do, Republic of Korea (37°01’47.3″N 127°11’15.4″E), isolated from soil in which pear trees was buried for disease control, collected on January 6, 2023, strain DUCC24276(NIBRFGC000510208).
Macro morphological characteristics (Fig. 9A–D)
The colony morphology was observed after 7 days of incubation in the dark condition at 25℃. In all media, strain NIBRFGC000510208 was fully grown to fill the plate. Colonies were fluffy, radially grown, gray colored, white clumped mycelia form at the edge. Subsequently, dark brown pigmentation resembling melanin developed, and a dense dot pattern was observed on the reverse side.
Microscopic morphological characteristics (Fig. 9E–H)
Hyphae were hyaline, thick, and 6.7–9.7 μm wide. Bulbils were observed after one month of incubation, were light-brown colored, consisted of densely thick hyphae, and were 100.01–105.53 × 76.64–86.44 μ m. Initially, the hook of the bulbil was produced in a tangled form at the end of the hyphae. Afterward, the bulbil hook formed tightly coiled hyphae. Cells that composed the bulbils were swollen, spherical, thickwalled, and 9.2–11.67 × 9.65–12.75 μm. Teleomorph was not observed.
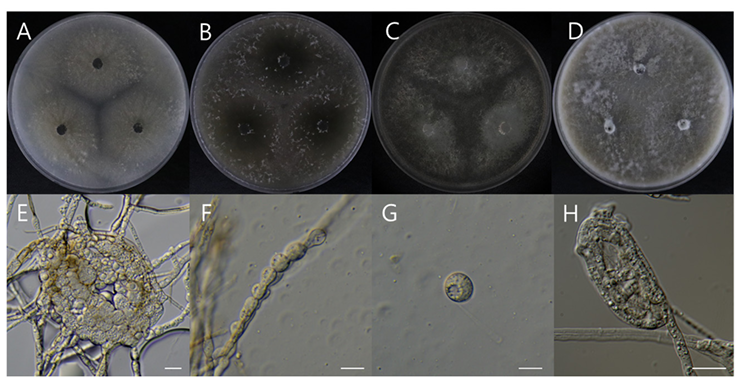
Fig. 9. Morphological characteristics of Papulaspora equi NIBRFGC000510208 incubated at 25 ℃ for 7 days. (A) PDA; (B) MEA; (C) CZA; (D) OA; (E) Bulbils; (F, G) Cells that compose the bulbils on OA; (H) Hook of bulbil on PDA. Scale bar = 10 μm. PDA: Potato Dextrose Agar; MEA: Malt Extract Agar; CYA: Czapek Yeast extract Agar; OA: Oatmeal Agar.
Molecular characteristics (Fig. 10)
The ITS and LSU rDNA sequences of strain NIBRFGC000510208 were used for phylogenetic analysis. The size of ITS and LSU rDNA sequences was 523 bp and 897 bp, respectively. These two sequences were concatenated and aligned with sequences of other Papulaspora species, resulting in a final sequence of 1,071 bp including gaps. Sequence similarity between strain NIBRFGC000510208 and P. equi CBS 573.89 (type strain) and CBS 128690 was 99.9%. It is higher than the accepted similarity threshold 90.0% in the clustering of strains for this species in MycoBank. As a result of constructing an ML phylogenetic tree based on the 1,071 bp concatenated sequence, strain NIBRFGC000510208 formed the same clade with P. equi CBS 573.89 (type strain) and CBS 128690
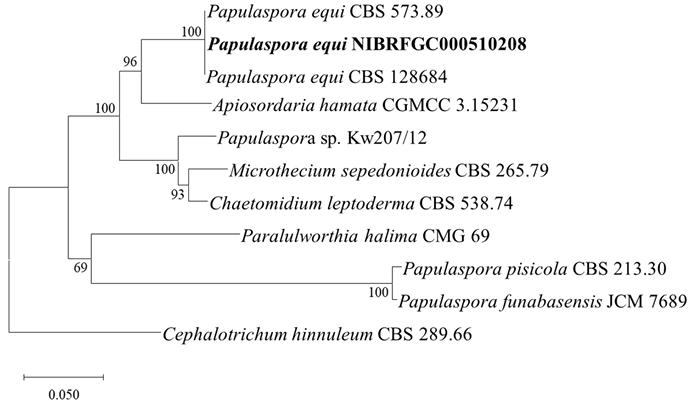
Fig. 10. The phylogenetic tree based on the concatenated nucleotide sequences of the ITS and LSU rDNA using the ML method with the Kimura 2-parameter model. The strain identified in this study is indicated in bold. The number of nodes represents the reliability value through 1,000 bootstrap replicates. Bootstrap values whose node reliability was less than 50 were removed. Cephalotrichum hinnuleum CBS 289.66 was used as an outgroup. ITS: internal transcribed spacer; LSU rDNA: 28S ribosomal RNA gene; ML: maximum likelihood.
Notes
The genus Papulaspora has been isolated from various objects, including soil, plants, and animals, but Papulaspora equi is a pathogenic fungus associated with the eyes. It is a potential zoonotic pathogen isolated from the infected eyes of horses and animals. Morphological characteristics of P. equi include the bulbils with an irregularly lobed surface and two sizes of hyphae; narrow hyphae 1–2.5 μm wide and broad hyphae 8 μm wide [38]. Additionally, P. equi is a rare causative agent of fungal keratomycosis in humans [39]. Strain NIBRFGC000510208 had morphologically similar characteristics to P. equi, such as thick hyphae and bulbils composed of thick cells. In phylogenetic analysis using the combined ITS and LSU rDNA sequences, strain NIBRFGC000510208 formed the same clade as that with the type strains P. equi CBS 573.89 and P. equi CBS 128690. Based on the results of morphological and molecular analysis, strain NIBRFGC000510208 was identified as Papulaspora equi, a species not previously recorded in Korea.
Strains examined: Seogwipo-si, Jeju Island, Republic of Korea (33°20’14.1″N 126°37’25.6″E), isolated from the hyposphere soil of Wrinkled mushroom, collected on June 19, 2023, strain DUCC15606(NIBRFGC000512619).
Macro morphological characteristics (Fig. 11A–D)
The colony morphology was observed after 7 days of incubation in the dark condition at 25℃. Colonies were circular, raised, and fluffy and had filiform margins. On CYA, growth reached approximately 30.53 mm, while on MEA, it grew slowly, reaching about 28.47 mm. On PDA and OA, the colony had a purple center, and on the reverse side, it was yellowish white-colored. On OA, synnemata formed within 7 days of culture, while in other media (PDA, MEA, CYA), it took 14 days.
Microscopic morphological characteristics (Fig. 11E–H)
Synnemata were formed by clusters of hyphae-gregarious, thick at the base and narrowing toward the terminal end, flat at the terminal end, 4.1–6.8 mm in length, 0.6–1 mm diam in base, and 0.4–0.6 mm diam in apex. Conidia are formed at the terminal end and margin. Vegetative hyphae were septate, hyaline, branched, smooth walled, 1.8–2.1 diam. Conidiophores were cylindrical, solitary, smooth walled, erect, or slightly curved, 6.5–7.2 × 2.1–2.4 μm. Phialides had occurred from vegetative hyphae or conidiophores and were ellipsoidal, cylindrical, and sometimes awl-shaped, and 11.28–18.03 × 1.55–2.3 μm. Conidia were single celled, hyaline, smooth walled, ovoid to ellipsoidal, continuously produced from phialides, and 3.24–3.52 × 2.62–2.84 μm. Teleomorph was not observed.
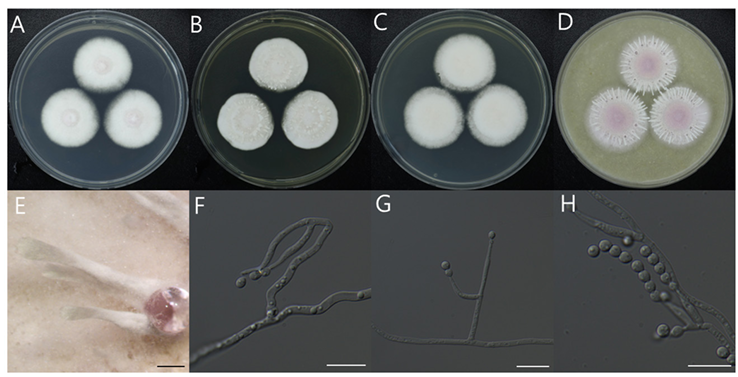
Fig. 11. Morphological characteristics of Paecilomyces purpureus NIBRFGC000512619 incubated at 25 ℃ for 7 days. (A) PDA; (B) MEA; (C) CYA; (D) OA; (E) synnemata; (F–H) Conidiophore, Conidiogenous cell and conidia; Scale bar (E = 1mm; F–H =10 μm). PDA: Potato Dextrose Agar; MEA: Malt Extract Agar; CYA: Czapek Yeast extract Agar; OA: Oatmeal Agar.
Molecular characteristics (Fig. 12)
The size of the ITS sequence was 491 bp. As a result of aligning sequence with the reference sequences of other Paecilomyces species, a final sequence of 506 bp including gaps was obtained. The ITS sequence of strain NIBRFGC000512619 was used for phylogenetic analysis. As a result of constructing an ML phylogenetic tree based on the 506 bp ITS sequences, strain NIBRFGC000510208 formed the same clade as that with P. purpureus GZDXIFR-GZSL-5.
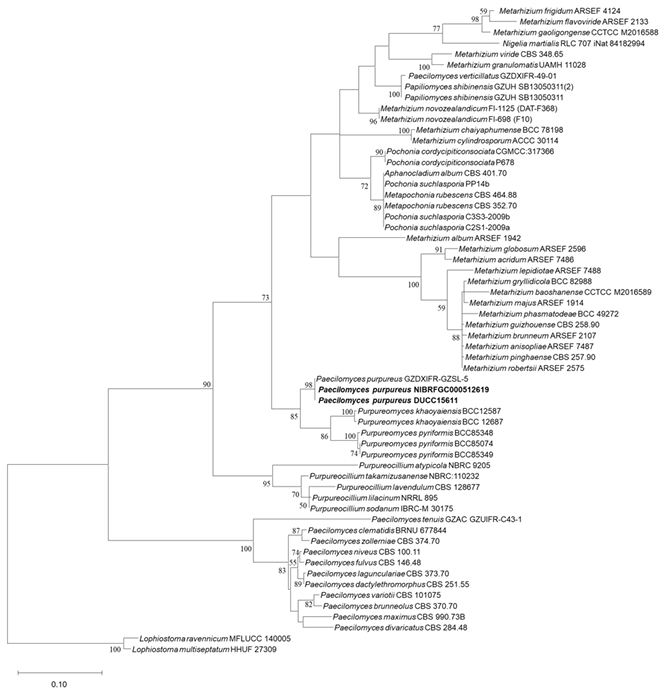
Fig. 12. The phylogenetic tree based on the nucleotide sequences of the ITS using the ML method with the Kimura 2-parameter model. Strains identified in this study are indicated in bold. The number of nodes represents the reliability value through 1,000 bootstrap replicates. Bootstrap values whose node reliability was less than 50 were removed. Lophiostoma ravennicum MFLUCC140005 and Lophiostoma multiseptatum HHUF 27309 were used as outgroups. ITS: internal transcribed spacer; ML: maximum likelihood.
Notes
Some members of the genus Paecilomyces are biostimulants of plant growth and crop yield [40]. They also produce secondary metabolites with herbicidal and cytotoxic activity [41,42]. The purple-red pigment produced by P. lilacinus TD16 is an important natural product for various applications. This species has antimicrobial activity against a wide range of bacteria and fungi, which suggests potential applications in industry [43]. In the genus, some species can be pathogenic to humans. P. lilacinus and P. variotii can mainly cause infections in immunocompromised individuals [44]. Morphologically, P. purpureus exhibited purple colonies; synnemata formed after long-term culture; phialides with a swollen basal portion or awl-shaped structure occurring directly or arising as 2–3 phialides from conidiophores; and single-celled, subglobose to fusiform conidia [45]. Strain NIBRFGC000512619 exhibited the morphological characteristics of P. purpureus, including purple synnemata, phialides with a swollen basal portion or awl-shaped structure occurring directly or arising as 2–3 phialides from conidiophores. In the phylogenetic analysis based on ITS rDNA sequences, strain NIBRFGC000512619 clustered with the same clade as P. purpureus GZDXIFRGZSL-5. Based on morphological and molecular analyses, strain NIBRFGC000512619 was identified as Paecilomyces purpureus, which has not been previously recorded in Korea.
The authors declare no conflicts of interest.
This work was supported by a grant from the National Institute of Biological Resources (NIBR), funded by the Ministry of Environment (MOE) of the Republic of Korea. The Department of Microbiology was supported through the Research Focused-Department Promotion & Interdisciplinary Convergence Research Project as a part of the Support Program for University Development for Dankook University in 2024.
1. Zhang N, Castlebury LA, Miller AN, Huhndorf SM, Schoch CL, Seifert KA, Rossman AY, Rogers JD, Kohlmeyer J, Volkmann-Kohlmeyer B, et al. An overview of the systematics of the Sordariomycetes based on a four-gene phylogeny. Mycologia 2006;98:1076-87. [DOI]
2. Hyde KD, Norphanphoun C, Maharachchikumbura SSN, Bhat DJ, Jones EBG, Bundhun D, Chen YJ, Bao DF, Boonmee S, Calabon MS, et al. Refined families of Sordariomycetes. Mycosphere 2020; 11:305-1059. [DOI]
3. Yasanthika WAE, Wanasinghe DN, Karunarathna SC, Bhat DJ, Samarakoon SMBC, Ren GC, Monkai J, Mortimer PE, Hyde KD. Two new Sordariomycetes records from forest soils in Thailand. Asian J Mycol 2020;3:456-71. [DOI]
4. Minahan NT, Chen CH, Shen WC, Lu TP, Kallawicha K, Tsai KH, Guo YL. Fungal spore richness in school classrooms is related to surrounding forest in a season-dependent manner. Microb Ecol 2022; 84:351-62. [DOI]
5. Luo ZL, Hyde KD, Liu JK, Maharachchikumbura SSN, Jeewon R, Bao DF, Bhat DJ, Lin CG, Li WL, Yang J, et al. Freshwater Sordariomycetes. Fungal Divers 2019;99:451-660. [DOI]
6. Qu J, Zou X, Cao W, Xu Z, Liang Z. Two new species of Hirsutella (Ophiocordycipitaceae, Sordariomycetes) that are parasitic on lepidopteran insects from China. MycoKeys 2021;82:81-96. [DOI]
7. de Silva DD, Groenewald JZ, Crous PW, Ades PK, Nasruddin A, Mongkolporn O, Taylor PWJ. Identification, prevalence and pathogenicity of Colletotrichum species causing anthracnose of Capsicum annuum in Asia. IMA Fungus 2019;10:8. [DOI]
8. Jaklitsch WM, Voglmayr H. Phylogenetic relationships of five genera of Xylariales and Rosasphaeria gen. nov (Hypocreales). Fungal Divers 2012;52:75-98. [DOI]
9. Guppy KH, Thomas C, Thomas K, Anderson D. Cerebral fungal infections in the immunocompromised host: a literature review and a new pathogen-Chaetomium atrobrunneum: case report. Neurosurgery 1998;43:1463-8. [DOI]
10. Castle A, Speranzini D, Rghei N, Alm G, Rinker D, Bissett J. Morphological and molecular identification of Trichoderma isolates on North American mushroom farms. Appl Environ Microbiol 1998;64:133-7. [DOI]
11. Sood M, Kapoor D, Kumar V, Sheteiwy MS, Ramakrishnan M, Landi M, Araniti F, Sharma A. Trichoderma: The “secrets” of a multitalented biocontrol agent. Plants 2020;9:762. [DOI]
12. Řehulka J, Kubátová A, Hubka V. Cephalotheca sulfurea (Ascomycota Sordariomycetes) a new fungal pathogen of the farmed rainbow trout Oncorhynchus mykiss. J Fish Dis 2016;39:1413-9. [DOI]
13. Dong C, Guo S, Wang W, Liu X. Cordyceps industry in China. Mycology 2015;6:121-9. [DOI]
14. Lee J. Characteristics of Korea’s climate. Soil Fertil 2000;1:84-93.
15. Tamura K, Stecher G, Kumar S. MEGA11: molecular evolutionary genetics analysis version 11. Mol Biol Evol 2021;38:3022-7. [DOI]
16. Cho A. Constructing phylogenetic trees using maximum likelihood [undergraduate thesis]. Claremont: Scripps College; 2012. Scripps Senior Theses:46.
17. Kimura M. A simple method for estimating evolutionary rate of base substitutions through comparative studies of nucleotide sequences. J Mol Evol 1980;16:111-20. [DOI]
18. Felsenstein J. Distance methods for inferring phylogenies: a justification. Evolution 1984;38:16-24. [DOI]
19. White TJ, Bruns TD, Lee SB, Taylor JW. Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics. In: Innis MA, Gelfand DH, Sninsky JJ, editors. PCR protocols: A guide to methods and applications. San Diego: Academic Press; 1990. p. 315-22. [DOI]
20. Cubeta MA, Echandi E, Abernethy T, Vilgalys R. Characterization of anastomosis groups of binucleate Rhizoctonia species using restriction analysis of an amplified ribosomal RNA gene. Phytopathology 1991;81:1395-400. [DOI]
21. Vilgalys R, Hester M. Rapid genetic identification and mapping of enzymatically amplified ribosomal DNA from several Cryptococcus species. J Bacteriol 1990;172:4238-46. [DOI]
22. Evidente A, Ricciardiello G, Andolfi A, Sabatini MA, Ganassi S, Altomare C, Favilla M, Melck D. Citrantifidiene and citrantifidiol: bioactive metabolites produced by Trichoderma citrinoviride with potential antifeedant activity toward aphids. J Agric Food Chem 2008;56:3569-73. [DOI]
23. Glass NL, Donaldson GC. Development of primer sets designed for use with the PCR to amplify conserved genes from filamentous ascomycetes. Appl Environ Microbiol 1995;61:1323-30. [DOI]
24. Liu YJ, Whelen S, Hall BD. Phylogenetic relationships among ascomycetes: evidence from an RNA polymerase II subunit. Mol Biol Evol 1999;16:1799-808. [DOI]
25. Subramanian CV, Lodha BC. Four new coprophilous Hyphomycetes. Antonie Van Leeuwenhoek 1964;30:317-30. [DOI]
26. Barrera VA, Martin ME, Aulicino M, Martínez S, Chiessa G, Saparrat MCN, Gasoni AL. Carbon-substrate utilization profiles by Cladorrhinum (Ascomycota). Rev Argent Microbiol 2019;51:302-6. [DOI]
27. Gams W, Klamer M, O’Donnell K. Fusarium miscanthi sp. nov. from Miscanthus litter. Mycologia 1999;91:263-8. [DOI]
28. Scauflaire J, Gourgue M, Foucart G, Renard F, Vandeputte F, Munaut F. Fusarium miscanthi and other Fusarium species as causal agents of Miscanthus× giganteus rhizome rot. Eur J Plant Pathol 2013; 137:1-3. [DOI]
29. Shang G, Yu H, Yang J, Zeng Z, Hu Z. First report of Fusarium miscanthi causing ear rot on maize in China. Plant Dis 2021;105:1565. [DOI]
30. Castellani A. A preliminary report on two pathogenic fungi: Trichophyton dankaliense n. sp.; Sporotrichum anglicum n. sp. J Trop Med Hyg 1937;40:313-8.
31. Deshmukh SK, Verekar SA. Isolation of keratinophilic fungi from selected soils of Sanjay Gandhi National Park, Mumbai (India). J Mycol Med 2014;24:319-27. [DOI]
32. Jambholkar S, Yadav S. Analysis of enzymatic activity of keratinophilic fungi. J Survey Fish Sci 2023;10:3268-73.
33. Amagata T, Minoura K, Numata A. Gymnastatins F−H, cytostatic metabolites from the sponge-derived fungus Gymnascella dankaliensis. J Nat Prod 2006;69:1384-8. [DOI]
34. Belcher BP, Machicao PA, Tong B, Ho E, Friedli J, So B, Bui H, Isobe Y, Maimone TJ, Nomura DK. Chemoproteomic profiling reveals that anti‐cancer natural product dankastatin B covalently targets mitochondrial VDAC3. Chembiochem 2023;24:e202300111. [DOI]
35. Sandoval-Denis M, Gené J, Sutton DA, Cano-Lira JF, De Hoog GS, Decock CA, Wiederhold NP, Guarro J. Redefining Microascus, Scopulariopsis and allied genera. Persoonia 2016;36:136. [DOI]
36. Mhmoud NA, Siddig EE, Nyuykonge B, Bakhiet SM, van de Sande WW, Fahal AH. Mycetoma caused by Microascus gracilis: a novel agent of human eumycetoma in Sudan. Trans R Soc Trop Med Hyg 2021;115:426-30. [DOI]
37. Ding Y, Steed LL, Batalis N. First reported case of disseminated Microascus gracilis infection in a lung transplant patient. IDCases 2020;22:e00984. [DOI]
38. Shadomy HJ, Dixon DM. A new Papulaspora species from the infected eye of a horse: Papulaspora equi sp. nov. Mycopathologia 1989;106:35-9. [DOI]
39. Selvin SST, Korah SMG, Michael JS, Raj PM, Jacob P. Series of five cases of Papulaspora equi keratomycosis. Cornea 2014;33:640-3. [DOI]
40. Moreno-Gavíra A, Diánez F, Sánchez-Montesinos B, Santos M. Paecilomyces variotii as a plant-growth promoter in horticulture. Agronomy 2020;10:597. [DOI]
41. Nakajima M, Itoi K, Takamatsu Y, Sato S, Furukawa Y, Furuya K, Honma T, Kadotani J, Kozasa M, Haneishi T. Cornexistin: a new fungal metabolite with herbicidal activity. J Antibiot 1991;44:1065-72. [DOI]
42. Nam KS, Jo YS, Kim YH, Hyun JW, Kim HW. Cytotoxic activities of acetoxyscirpenediol and ergosterol peroxide from Paecilomyces tenuipes. Life Sci 2001;69:229-37. [DOI]
43. Yu X, Xing X, Dong Q, Shi K, Yan R. Separation, quantification and characterisation of the pigment produced by Paecilomyces lilacinus TD16. Nat Prod Res 2022;36:5053-7. [DOI]
44. Pastor FJ, Guarro J. Clinical manifestations, treatment and outcome of Paecilomyces lilacinus infections. Clin Microbiol Infect 2006;12:948-60. [DOI]
45. Liang Z, Han Y, Liang J, Zou X. Paecilomyces purpureus sp. nov., a new entomogenous fungal species from China. Mycotaxon 2007;101:271-8.