Mohammad Hamizan Azmi1, Seong-Keun Lim1, Gwang-Jae Lim1, Jin-Sil Choi1, Jun-Woo Choi1, MinGyu Kim1, Seung-Yeol Lee1,2*, and Hee-Young Jung1,2
1Department of Plant Medicine, Kyungpook National University, Daegu 41566, Korea
2Institute of Plant Medicine, Kyungpook National University, Daegu 41566, Korea
*Correspondence to leesy1123@knu.ac.kr
Korean Journal of Mycology (Kor J Mycol) 2024 December, Volume 52, Issue 4, pages 391-398.
https://doi.org/10.4489/kjm.520417
Received on December 18, 2024, Revised on December 20, 2024, Accepted on December 20, 2024, Published on Dec 30, 2024.
Copyright © The Korean Society of Mycology.
This is an Open Access article which is freely available under the Creative Commons Attribution-NonCommercial 4.0 International License (CC BY-NC) (https://creativecommons.org/licenses/by-nc/4.0/).
The fungal strain KNUF-21-015 was isolated from soil in Palgongsan, Daegu-si, Gyeongbuk province in Korea, and identified as a previously unreported species within the family Diatrypaceae. Observations of conidiogenous cells, pycnidia, and conidia were conducted, and morphological differences between strain KNUF-21-015 and closely related species were compared. As the asexual morph of this species had not been documented previously, phylogenetic analyses were performed using concatenated nucleotide sequences from the internal transcribed spacer (ITS) regions and β-tubulin (TUB) gene to elucidate its identity and evolutionary relationships. The results placed the strain within the genus Diatrype, with high sequence similarities of 99.8 and 99.0% to D. rubi GMB0429T for the ITS regions and TUB gene, respectively. Phylogenetic and morphological evidence collectively support the identification of the strain KNUF-21-015 as the asexual morph of D. rubi. To the best of our knowledge, this is the first report of D. rubi and its anamorph in Korea.
Diatrypaceae, Diatrype rubi, Morphology, Phylogenetic analyses, Soil
The family Diatrypaceae, which belongs to the order Xylariales, class Sordariomycetes, and phylum Ascomycota, comprises 28 genera and is widely distributed [1–3]. Members of this family are commonly found on decaying wood and the bark of diverse plant species, contributing to their ubiquitous presence across diverse ecosystems [3]. The genus Diatrype, a key member of the Diatrypaceae family, has a documented distribution spanning Asia, Europe, North America, Oceania, and South Africa [4]. According to the Mycobank database (https://mycobank.org) approximately 401 species of Diatrype are recognized, though only two species, D. stigma and D. disciformis, have been previously reported in Korea [5–7].
The taxonomic framework of Diatrypaceae has historically been unstable, with numerous new genera emerging through the integration of morphological traits and multi-locus phylogenetic analyses [3]. Traditionally, the classification of Diatrypaceae species has relied heavily on stromatal characteristics. However, this approach has led to significant confusion, resulting in polyphyletic genera and frequent species reassignments [3]. The genus Diatrype was first established by Fries in 1894 under the family Diatrypaceae, with D. disciformis as the type species [8]. The asexual morph of this genus is described as libertella-like, characterized by pycnidial conidiomata, while its sexual morph features stromata that are widely effused or verrucose, with eight-spored, long-stalked asci, and hyaline or brownish, allantoid ascospores [2,8,9].
This study aimed to isolate fungi from domestic soil samples in Korea and characterize them based on morphological and phylogenetic traits. The goal is to document potential endemic species, preserve domestic fungal resources, and contribute to a more comprehensive understanding of the fungal biodiversity in Korea.
Soil samples for fungal isolation were collected from Mt. Palgongsan, Daegu-si, Gyeongbuk province, Korea (35°59’33.9″ N, 128°41’12.7″ E). Fungi were isolated using the serial dilution method, as described in a previous study [10]. Single colonies were then transferred to fresh potato dextrose agar (PDA; Difco, Detroit, MI, USA) plates and incubated at 25℃. The fungal strain KNUF-21-015 was selected for molecular, cultural, and morphological analyses. This isolate is preserved at the National Institute of Biological Resources (NIBR) under the accession number NIBRFGC000509191.
The strain KNUF-21-015 was cultured at 25℃ on PDA and potato carrot agar (PCA: potato, 20 g; carrot, 20 g; agar, 20 g; distilled H2 O, 1,000 mL) for morphological and cultural characterization. Cultures were maintained in the dark, and after 7 days, characteristics such as the size, color, and shape of the mycelium, as well as the conidiogenous cells and conidia, were observed. A stereoscopic microscope (Digital Micro Scope-M5; Siwon, Anyang, Korea) and a light microscope (BX-50; Olympus, Tokyo, Japan) were used to examine the morphological and cultural properties.
For phylogenetic analysis, genomic DNA from strain KNUF-21-015 was extracted using the HiGene Genomic DNA Prep Kit (Biofact, Daejeon, Korea) following the manufacturer’s instructions. The internal transcribed spacer (ITS) regions and β-tubulin (TUB) gene were amplified using the ITS1F/ITS4 and T1/ Bt2b primer pairs, respectively [11–13]. Successful amplification was confirmed through electrophoresis on 1.0% HP Agarose (BIOPURE, Cambridge, USA) gels. The amplified products were purified using ExoSAP-IT (Thermo Fisher Scientific, Waltham, MA, USA) and submitted to Macrogen (Seoul, Korea) for sequencing.
The sequences of strain KNUF-21-015 were analyzed for similarity using the Basic Local Alignment Search Tool (BLAST) against datasets in the National Center for Biotechnology Information (NCBI) database. Subsequently, several related sequences were retrieved from the database to conduct phylogenetic analyses (Table 1). Phylogenetic trees were constructed using the concatenated sequences of the ITS regions and TUB gene, employing the neighbor-joining (NJ) method in MEGA X [14,15]. Evolutionary distance matrices for the NJ analysis were calculated using Kimura’s two-parameter model, with bootstrap values based on 1,000 replications [16].
Table 1. List of species used in phylogenetic analyses along with their GenBank accession numbers
| Species name | Strain | GenBank accession number | |
|---|---|---|---|
| ITS | TUB | ||
| Diatrype betulaceicola | FCATAS 2725T | OM040386 | OM240966 |
| Diatrype betulaceicola | FCATAS 2726 | OM040387 | OM240967 |
| Diatrype betulae | CFCC 52416T | MW632943 | MW656391 |
| Diatrype bullata | UCDDCh400 | DQ006946 | DQ007002 |
| Diatrype camelliae-japonicae | GMB0427T | OP935172 | OP938734 |
| Diatrype camelliae-japonicae | GMB0428 | OP935173 | OP938735 |
| Diatrype castaneicola | CFCC55425T | MW632941 | MW656389 |
| Diatrype castaneicola | CFCC52426 | MW632942 | MW656390 |
| Diatrype disciformis | GB 5815 | KR605644 | KY352434 |
| Diatrype lancangensis | GMB0045T | MW797113 | MW814885 |
| Diatrype lancangensis | GMB0046 | MW797114 | MW814886 |
| Diatrype larissae | FCATAS 2723T | OM040384 | OM240964 |
| Diatrype larissae | FCATAS 2724 | OM040385 | OM240965 |
| Diatrype lijiangensis | MFLU 19-0717T | MK852582 | MK852583 |
| Diatrype quercicola | CFCC 52418T | MW632938 | MW656386 |
| Diatrype quercicola | CFCC 52419 | MW632939 | MW656387 |
| Diatrype rubi | GMB0429T | OP935182 | OP938740 |
| Diatrype rubi | KNUF-21-015 | OR727457 | OR729842 |
| Diatrype stigma | DCash200 | GQ293947 | GQ294003 |
| Diatrype stigma | Kern307 | OP038055 | OP079884 |
| Xylaria hypoxylon | CBS 122620 | KY610407 | KX271279 |
ITS: internal transcribed spacer regions; TUB: β-tubulin gene.
T Type strain. Strain used in this research is highlighted in bold.
After 7 d of incubation on PDA at 25℃, strain KNUF-21-015 formed colonies with a 90 mm diameter. The obverse side of the colony was white and dense, gradually thinning toward the edge, with a rough margin. The reverse side exhibited a sepia tone extending from the center to the margin, lacked pigmentation on the medium, and appeared filamentous (Fig. 1A). Under the same conditions, the strain grown on PCA formed smaller colonies, measuring 37–38 mm in diameter. The obverse side of the colony was white and filamentous, with a rough margin, while the reverse ranged from yellowish to light brown, also without media pigmentation (Fig. 1B). Orange color pycnidia were observed on PCA media (Fig. 1C). Conidiogenous cells derived from the pycnidia were cylindrical, straight, discrete or integrated, hyaline, and produced conidia at the apex (Fig. 1D and E). The conidia were hyaline, straight to slightly curved, allantoid, and aseptate, measuring 10.8–23.9 × 1.0–2.0 µm (avg. = 19.5 × 1.5 µm, n = 50) (Fig. 1F).
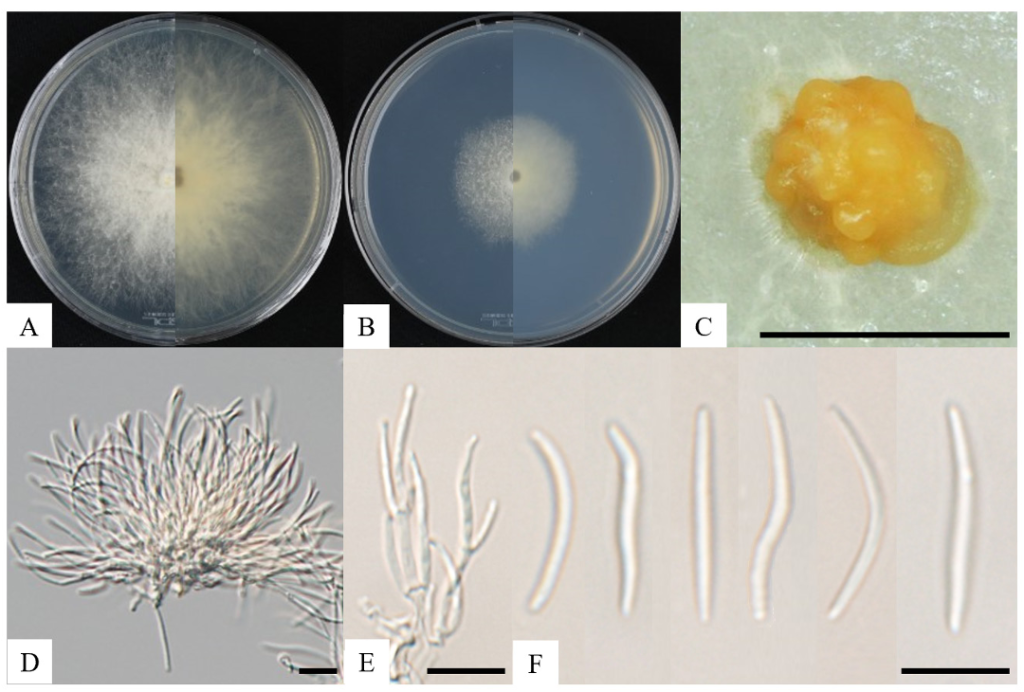
Fig. 1. Cultural and morphological characteristics of Diatrype rubi KNUF-21-015. Cultures were grown at 25℃ for 7 days. A and B: Obverse and reverse view of the colony on PDA and PCA; C: Pycnidia on PCA; D: Mass of conidiogenous cell and conidia; E: Conidia with conidiogenous cell; F: Conidia. Scale bars: C = 50 µm, D–F = 10 µm.
Molecular identification of the isolated fungal strain KNUF-21-015 involved amplifying its ITS regions and TUB gene, yielding sequences of 602 and 707 bp, respectively. The ITS regions exhibited 99.8% similarity to D. rubi GMB0429T, compared to 99.0% with D. camelliae-japonicae GMB0427T. Similarly, the TUB gene showed 99.0% similarity with D. rubi GMB0429T, compared to 97.3% with D. camellia e-japonicae GMB0427T. The NJ phylogenetic tree constructed from the concatenated sequences of the ITS regions and TUB gene clustered strain KNUF-21-015 with D. rubi GMB0429T (Fig. 2). Combined morphological, cultural, and phylogenetic analyses identified the strain as D. rubi (Table 2).

Fig. 2. Neighbor-joining phylogenetic tree based on a combined dataset of partial sequences internal transcribed spacer (ITS) regions and β-tubulin (TUB) gene showing the phylogenetic position of the strain KNUF-22-015 among Diatrype species. Bootstrap values greater than 90% (percentage of 1,000 replications) are shown at branching points. The strain isolated in this study is in bold and red. The tree was rooted using Xylaria hypoxylon CBS 122620 as an out-group. Bar = 0.02 substitutions per nucleotide position.
Table 2. Morphological characteristics of Diatrype rubi KNUF-21-015 compared to closely related Diatrype species
| Characteristics | Diatrype rubi KNUF-21-015a | Diatrype rubi GMB0429Tb | Diatrype castaneicola CFCC52425Tc | |
|---|---|---|---|---|
| Colony on PDA | Color | White, reverse sepia, no pigmentation on media | White to light yellow, reverse white at the margin, mauve to sepia at the center | White, no pigmentation on media |
| Shape | Dense, thinning toward the edge, rough margin, filamentous | Dense, thinning toward the edge, rough margin | Dense, pycnidia distributed irregularly with yellow cream conidial drops | |
| Conidiogenous cell | Cylindrical, straight, discrete or integrated, hyaline, producing conidia at the apex | N/A | Cylindrical, mostly straight, discrete or integrated, hyaline, unicellular, with wide base producing conidia at the apex | |
| Conidia | Size | 10.8–23.9 × 1.0–2.0 µm | N/A | 4.0–6.0 × 1.0–1.5 µm |
| Shape | Hyaline, slightly curved, allantoid, aseptate | Hyaline, elongate-allantoid, slightly curved, smooth, aseptate, multi-guttulate |
PDA: potato dextrose agar; N/A: not available in the described paper.
aFungal strain used in this study; b Source of description [1]; c Source of description[22]; T Type strain.
The ambiguity in Diatrype taxonomy, largely driven by the reliance on stromatal characteristics, has resulted in polyphyletic genera and inconsistent species classifications. To resolve these issues, we employed multi-locus phylogenetic analyses, focusing on the ITS regions and TUB gene [4]. These analyses provided a more accurate determination of Diatrype species and assisted in identifying their plant pathogenicity. Members of the genus Diatrype are known to cause grapevine trunk diseases (GTDs) by attacking woody tissues and inducing trunk rots [17]. While several Diatrype species are recognized as pathogenic to grapevines [4,18], specific research on the pathogenicity of D. rubi is lacking. Notably, this species has also been observed to grow saprophytically on its host, suggesting the need for further study to assess its pathogenic potential. Diatrype rubi was first reported in China [1], where it was isolated as a saprobe on the branch surface of its host Rubus corchorifolius. The species was first discovered in its sexual morph, while its asexual morph remained uncharacterized. Previous studies indicate that the asexual morphs of many fungi are commonly observed when strains are grown on PCA media [19–22]. Consistent with these findings, strain KNUF-21-015 was cultured on PCA, revealing pycnidia, conidiogenous cells, and conidia. As the anamorph of D. rubi had not been reported previously, the observed characteristics were compared to the anamorphs of closely related Diatrype species, such as D. castaneicola (Table 2). This study presents the first report of the species D. rubi in Korea, as well as the first documentation of its anamorph, increasing the number of Diatrype species reported in Korea to three: D. disciformis, D. stigma, and D. rubi [5–7]. Considering the high number of reported species worldwide, there is a high likelihood of more species of the genus Diatrype to be reported in the Korean peninsula. The identification of the anamorph not only enhances the taxonomic understanding of Diatrype species but also serves as a critical step in exploring their ecological roles and pathogenicity. These findings lay a strong foundation for future research into the biology, distribution, and pathogenicity of D. rubi and other Diatrype species.
No conflict of interests was reported or declared by the authors.
This research was performed with the support of the grant from the National Institute of Biological Resources (NIBR), funded by the Ministry of Environment (MOE) of the Republic of Korea (NIBR202102107).
1. Li QR, Long SH, Lin Y, Wu YP, Wu QZ, Hu HM, Shen XC, Zhang X, Wijayawardene NN, Kang JC, et al. Diversity, morphology, and molecular phylogeny of Diatrypaceae from southern China. Front Microbiol 2023;14:1140190. [DOI]
2. Ma HX, Yang ZE, Song ZK, Qu Z, Li Y, Zhu AH. Taxonomic and phylogenetic contributions to Diatrypaceae from southeastern Tibet in China. Front Microbiol 2023;14:1073548. [DOI]
3. Habib K, Zhou X, Zeng W, Zhang X, Hu H, Wu Q, Liu L, Lin Y, Shen X, Kang J, et al. Stromatolinea, a new diatrypaceous fungal genus (Ascomycota, Sordariomycetes, Xylariales, Diatrypaceae) from China. MycoKeys 2024;108:197-225. [DOI]
4. Yang Z, Zhang B, Qu Z, Song Z, Pan X, Zhao C, Ma H. Two new species of Diatrype (Xylariales, Ascomycota) with polysporous asci from China. Diversity 2022;14:149. [DOI]
5. National Institute of Biological Resources (NIBR). National list species of Korea [Internet]. Incheon: NIBR; 2023 [cited 2024 Nov 11]. Available from: https://species.nibr.go.kr.
6. Bak WC, Lee BH, Yoon KH, Ka KH, Choi JS, Lee TS. Observation of anamorph (Libertella sp.) and teleomorph (Diatrype stigma) of D. stigma affecting bed-log of oak-mushroom in Korea. Kor J Mycol 2000;28:38-40.
7. Lee SW, Choi J, Won HY, Lee YS, Yu D, Han A, Lee HY, Lee HS, Eo JK. The funga of higher fungi of Mt. Jeombong in Korea: a survey of Mongolian oak forest in 2017. Geo Data 2023;5:40-8. [DOI]
8. Rappaz F. Taxonomie et nomenclature des Diatrypacees à asques octospores. Mycol Helv 1987;2:285-648.
9. Thiyagaraja V, Senanayake IC, Wanasinghe DN, Karunarathna SC, Worthy FR, To-Anun C. Phylogenetic and morphological appraisal of Diatrype lijiangensis sp. nov. (Diatrypaceae, Xylariales) from China. Asian J Mycol 2019;2:198-208. [DOI]
10. Kim TG, Ten LN, Hong SM, Lim SK, Lee SY, Jung HY. First report of Hamigera ingelheimensis isolated from Cheoltan mountain in Korea. Kor J Mycol 2024;52:155-63.
11. White TJ, Bruns TD, Lee SB, Taylor JW. Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics. In: Innis MA, Gelfand DH, Sninsky JJ, editors. PCR protocols: a guide to methods and applications. San Diego: Academic Press; 1990. p. 315-22. [DOI]
12. Glass NL, Donaldson GC. Development of primer sets designed for use with the PCR to amplify conserved genes from filamentous ascomycetes. Appl Environ Microbiol 1995;61:1323-30. [DOI]
13. O’Donnell K, Cigelnik E. Two divergent intragenomic rDNA ITS2 types within a monophyletic lineage of the fungus Fusarium are nonorthologous. Mol Phylogenet Evol 1997;7:103-16. [DOI]
14. Saitou N, Nei M. The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol Biol Evol 1987;4:406-25.
15. Kumar S, Stecher G, Li M, Knyaz C, Tamura K. MEGA X: molecular evolutionary genetics analysis across computing platforms. Mol Biol Evol 2018;35:1547-9. [DOI]
16. Kimura M. A simple method for estimating evolutionary rates of base substitutions through comparative studies of nucleotide sequences. J Mol Evol 1980;16:111-20. [DOI]
17. Mundy DC, Brown A, Jacobo F, Tennakoon K, Woolley RH, Vanga B, Tyson J, Johnston P, Ridgway HJ, Bulman S. Pathogenic fungi isolated in association with grapevine trunk diseases in New Zealand. New Zeal J Crop Hort Sci 2020;48:84-96. [DOI]
18. Pitt WM, Trouillas FP, Gubler WD, Savocchia S, Sosnowski MR. Pathogenicity of diatrypaceous fungi on grapevines in Australia. Plant Dis 2013;97:749-56. [DOI]
19. Al-Musa A, Al-Maqtoofi M, Alrubayae I, Altooma M. New species of Preussia from sedimentary cost in Basrah province, Iraq. Eur J Biol Biotechnol 2024;5:16-20. [DOI]
20. Alizadeh M, Safaie N, Shams-Bakhsh M, Mehrabadi M. Neoscytalidium novaehollandiae causes dieback on Pinus eldarica and its potential for infection of urban forest trees. Sci Rep 2022;12:9337. [DOI]
21. Ujat AH, Hattori Y, Masuya H, Fatin AHKF, Nakashima C. Diversity of caulicolous species of the genus Diaporthe on Prunus sensu lato in Japan. Plant Fung Res 2024;7:2-24.
22. Zhu H, Pan M, Wijayawardene NN, Jiang N, Ma R, Dai D, Tian C, Fan X. The hidden diversity of diatrypaceous fungi in China. Front Microbiol 2021;12:646262. [DOI]