Minseo Cho1, Yeonjae Yoo1, Sang Hyun Lee1, Dae Young Kwon1, Sun Lul Kwon2*, and Jae-Jin Kim1*
1Division of Environmental Science and Ecological Engineering, College of Life Sciences and Biotechnology, Korea University, Seoul 02841, Korea
2Division of Wood Engineering, National Institute of Forest Science, Seoul 02455, Korea
*Correspondence to sun-lul@korea.ac.kr, jae-jinkim@korea.ac.kr
Korean Journal of Mycology (Kor J Mycol) 2024 December, Volume 52, Issue 4, pages 399-411.
https://doi.org/10.4489/kjm.520418
Received on December 20, 2024, Revised on December 20, 2024, Accepted on December 20, 2024, Published on Dec 30, 2024.
Copyright © The Korean Society of Mycology.
This is an Open Access article which is freely available under the Creative Commons Attribution-NonCommercial 4.0 International License (CC BY-NC) (https://creativecommons.org/licenses/by-nc/4.0/).
Macrofungi are major decomposers in terrestrial ecosystems, acting as nutrient cyclers, wood decomposers, and plant symbionts. Despite their beneficial effects, the diversity of macrofungi is not as well understood as that of the flora and fauna. Mudeungsan National Park is one of the biggest National Parks in Southwestern Korea. We conducted surveys in Mudeungsan National Park from 2021 to 2023 to identify indigenous macrofungi. The collected specimens were identified based on morphological characteristics and molecular and phylogenetic analyses using the internal transcribed spacer (ITS), nuclear large subunit ribosomal DNA (nLSU), and translation elongation factor 1 gene (tef1) regions. Four species were identified as unrecorded in Korea: Anomoloma luteoalbum, Fibrodontia alba, Russula pallidula, and Schizocorticium parvisporum. In this study, we provide detailed morphological figures and phylogenetic trees to support the four unrecorded species in Mudeungsan National Park.
Basidiomycota, Indigenous fungi, Phylogeny, Taxonomy
Macrofungi, such as ectomycorrhizal, saprobic, parasitic, and wood-decaying fungi, are major players in terrestrial ecosystems. They contribute to nutrient cycling, wood decomposition, and plant growth through mycorrhizal interactions [1–4]. In addition, several macrofungi provide economic benefits owing to their medicinal and nutritional values [5]. Technological advancements in DNA-based analyses estimate that 11.7–13.2 million fungal species exist on Earth [6]. However, macrofungal species have been recently estimated to be 220,000–380,000 [7], following the result of 2.2–3.8 million fungal species in 2017 [8]. According to Hawksworth (2001), 10% of the fungal species are macrofungi [9]. Thus, the current estimate should be updated to 1.2–1.3 million species following Hawksworth’s calculations [9]. In Korea, 5,054 fungal species have been recorded until 2023, of which 2,188 have been identified as macrofungal species [10]. This indicates that macrofungal diversity in Korea is still limited compared to global diversity.
Mudeungsan National Park is located across Hwasun-gun and Gwangju Metropolitan City in Jeollanamdo province, Korea. Mudeungsan National Park was first established as a provincial park in 1972 and later upgraded to a National Park in 2012. Moreover, it was registered as a UNESCO Global Geopark in 2018, recognizing its biological and geological value. Several studies have identified the diversity of flora and fauna in Mudeungsan National Park [11–14], but data on macrofungal diversity are still limited [15].
Previous research in Mudeungsan National Park showed the possibility of high macrofungal diversity and argued for the need for further research [15]. This survey was conducted as part of a project by the National Institute of Biological Resources to gain a better understanding of macrofungal diversity in Mudeungsan National Park. In the present study, we identified four previously unrecorded species based on morphological and molecular analyses. This study provides micro- and macro-morphological descriptions of each species, along with phylogenetic trees, and reports them as new to Korea.
Specimens were collected from Mudeungsan National Park (35°03′06ʺ–35°12′59ʺ N, 126°53′41ʺ–127° 05′01ʺ E) between 2021 and 2023. After collection, the samples were dried at 55℃ for 72 h and stored in paper bags containing silica gel to avoid contamination. The specimens were subsequently deposited at the Korea University Culture Collection (KUC) and National Institute of Biological Resources (NIBR).
Genomic DNA was isolated from each specimen using the AccuPrep® Genomic DNA Extraction Kit (Bioneer, Daejeon, Korea) and subjected to PCR amplification. The nuclear ribosomal internal transcribed spacer (ITS) region was amplified using primer sets ITS1F/ITS4 and ITS5/ITS4 [16,17]. For nuclear large subunit ribosomal DNA (nLSU), the LR0R/LR5 primer set was used [18,19], and the translation elongation factor 1 (tef1) gene region was amplified using EF1-983F/EF1-1567R primers [20]. PCR products were purified using the AccuPrep® PCR Purification Kit (Bioneer), following the protocol provided by the manufacturer. The sequencing was performed by Bioneer and Cosmogenetech (Seoul, Korea). The obtained sequences were assembled, proofread, and edited using Geneious Prime 2023.2.1. (https://www. geneious.com). The sequences obtained in this study were submitted to GenBank and are listed in Table 1.
Table 1. List of studied specimens with GenBank accession numbers of ITS, nLSU, and tef1 sequences.
| Species name | Specimen voucher | Order | GenBank accession number | ||
|---|---|---|---|---|---|
| ITS | nLSU | tef1 | |||
| Anomoloma luteoalbum | KUC20210714-74 | Amy | PQ619322 | PQ619355 | – |
| KUC20230317-01 | Amy | PQ620178 | – | – | |
| Fibrodontia alba | KUC20210930-17 | Tre | PQ619331 | PQ619362 | – |
| Russula pallidula | KUC20220715-36 | Rus | PQ619349 | PQ619379 | – |
| Schizocorticium parvisporum | KUC20220414-13 | Hym | – | PQ619380 | PQ661210 |
| KUC20220414-22 | Hym | PQ619350 | PQ619381 | PQ661211 | |
ITS: internal transcribed spacer; nLSU: nuclear large subunit ribosomal DNA; tef1: translation elongation factor 1 gene; Amy: Amylocortiales; Tre: Trechisporales; Rus: Russulales; Hym: Hymenochaetales.
For the phylogenetic analysis, reference sequences of related species were retrieved from the NCBI GenBank database [21]. Sequence alignment was performed using MAFFT v. 7.490 [22], with manual adjustments for ambiguous regions. Concatenated sequences were prepared using Geneious Prime 2023.2.1. A phylogenetic tree was constructed using the maximum likelihood (ML) method in the CIPRES web portal [23]. ML analysis was performed using RAxML-HPC2 in XSEDE v.8.2.12, applying the GTR+G model and 1,000 bootstrap replicates [24]. Phylogenetic trees were edited using FigTree v. 1.4.4 [25] and Adobe Illustrator CS6 v. 25.2.0 (Adobe Systems Inc., San Jose, CA, USA).
Macroscopic images of the specimens were captured using a Sony α 6500 camera (Sony, Tokyo, Japan). Micromorphological characteristics were observed using an Olympus BX51 light microscope (Olympus, Tokyo, Japan), and pictures were taken using a DP20 microscope camera (Olympus). Tissues from each specimen were mounted in 5% KOH for microscopic observation. Color terminology adhered to the standards outlined in the Munsell Soil Color Book [26]. The abbreviations used are as follows: L = average spore length, W = average spore width, and Q = L/W ratio.
This study discovered four previously unrecorded species through multi-marker phylogenetic analyses (Figs. 1–4). An additional tef1 region was used for Schizocorticium parvisporum owing to the uncertainty of species identification using ITS+nLSU data [27]. The four previously unrecorded species belong to the orders Amylocorticiales, Hymenochaetales, Russulales, and Trechisporales (Table 1).
According to the phylogenetic analyses, all unrecorded species showed high sequence similarity to their reference sequences and formed distinct clades with high bootstrap values (Figs. 1–4). Two genera were new to Korea: Fibrodontia Parmasto and Schizocorticium Sheng H. Wu. Schizocorticium was previously classified as Skvortzoviella Jia Yu, Xue W. Wang, S.L. Liu, and L.W. Zhou, and is currently synonymized as Schizocorticium. Additionally, three species-Fibrodontia alba, Russula pallidula, and Schizocorticium parvisporum-are new to temperate regions above 30° latitude, and were previously reported in tropical to subtropical areas, such as Southern China and Taiwan [27–29].
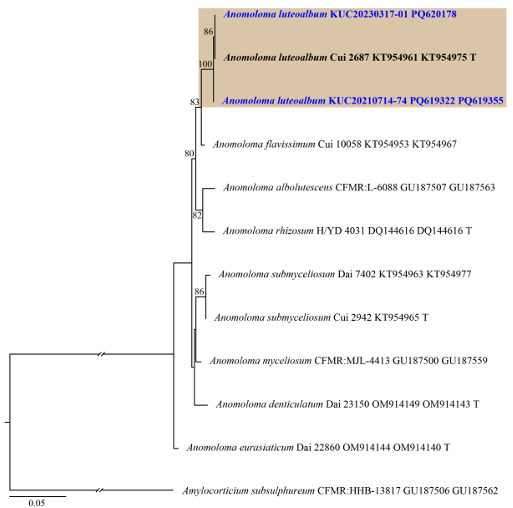
Fig. 1. ML tree based on the ITS and nLSU concatenated sequence datasets of the genus Anomoloma. The node numbers indicate the bootstrap support values above 70%. Newly generated sequences in this study are shown in blue and bold. Amylocorticium subsulphureum (CFMR:HHB-13817) was used as an outgroup. Type specimens are indicated with “T”. ML: maximum likelihood; ITS: internal transcribed spacer; nLSU: nuclear large subunit ribosomal DNA.
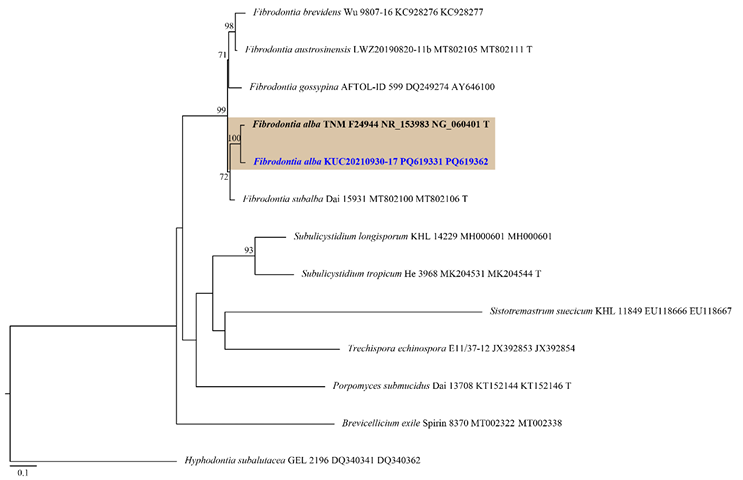
Fig. 2. ML tree based on the ITS and nLSU concatenated sequence datasets of the genus Fibrodontia and allied genera. The node numbers indicate the bootstrap support values above 70%. Newly generated sequence in this study is shown in blue and bold. Hyphodontia subalutacea (GEL 2196) was used as an outgroup. Type specimens are indicated with “T”. ML: maximum likelihood; ITS: internal transcribed spacer; nLSU: nuclear large subunit ribosomal DNA.
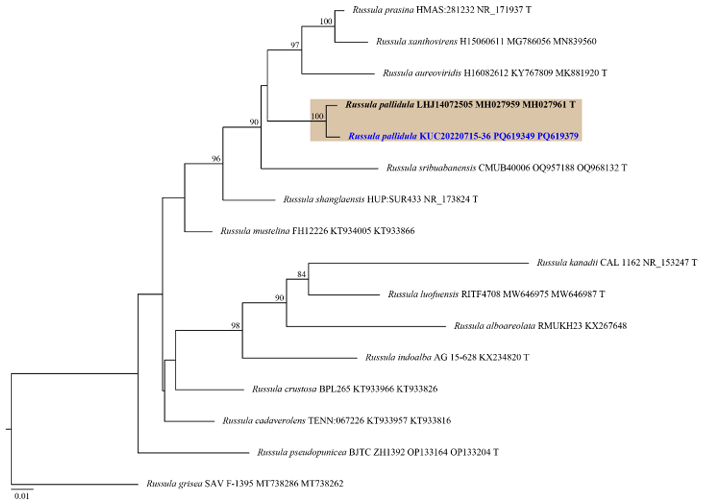
Fig. 3. ML tree based on the ITS and nLSU concatenated sequence datasets of the genus Russula. The node numbers indicate the bootstrap support values above 70%. Newly generated sequence in this study is shown in blue and bold. Russula grisea (SAV F-1395) was used as an outgroup. Type specimens are indicated with “T”. ML: maximum likelihood; ITS: internal transcribed spacer; nLSU: nuclear large subunit ribosomal DNA.
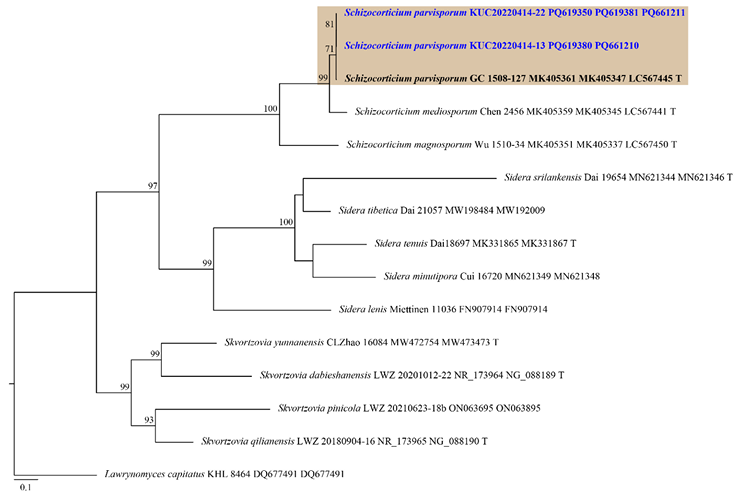
Fig. 4. ML tree based on the ITS, nLSU and tef1 concatenated sequence datasets of the genus Schizocorticium and allied genera. The node numbers indicate the bootstrap support values above 70%. Newly generated sequences in this study are shown in blue and bold. Lawrynomyces capitatus (KHL 8464) was used as an outgroup. Type specimens are indicated with “T”. ML: maximum likelihood; ITS: internal transcribed spacer; nLSU: nuclear large subunit ribosomal DNA; tef1: translation elongation factor 1 gene.
The finding that several macrofungi previously reported in tropical to subtropical areas are now found in temperate regions suggests that climate change affects macrofungal distribution. Because the life cycle of macrofungi is closely associated with trees, they are likely to be affected by changes in tree distribution due to climate change. Several studies have shown that macrofungal composition and diversity are affected by the composition and diversity of trees [30–32]. In addition, a recent study revealed that future climate and tree distribution changes affect the biodiversity of macrofungi [33]. Mudeungsan National Park is located in the Southern region of Korea. It is expected to enter a subtropical climate more quickly and to be more strongly impacted by climate change than other National Parks in Northern areas [34,35]. Therefore, further research is required to identify the long-term macrofungal diversity and discover new or unrecorded species in Mudeungsan National Park.
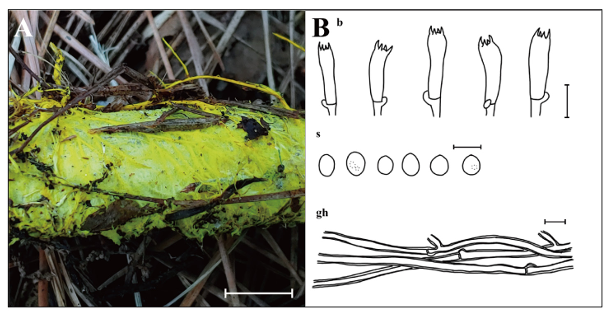
Fig. 5. Morphological characteristics of Anomoloma luteoalbum KUC20210714-74 and KUC2023031701. A: Basidiome; B: Microscopic structures, b: basidia; gh: generative hyphae; s: basidiospores (Scale bars: A = 1 cm, B = 10 μm, 5 μm for s).
MycoBank number: MB 821239
Basidiome resupinate, effused, soft when fresh, corky when dried. Margin irregular, abundant yellow (2.5Y, 8/8) rhizomorphs grow out from margin. Pores angular, 5–6 per mm, surface bright yellow (2.5Y, 8/8). Tubes soft, corky, concolorous with pore surface, up to 0.3 mm long. Subiculum yellow (2.5Y, 8/8) to pale brown (2.5Y, 8/4) when dry, up to 0.7 mm thick. Basidia with 4–spored, cylindrical to subclavate, 16.1–23.8 × (3.8–)4.1–7.3 μm, smooth, hyaline, thin-walled, with a basal clamp connection. Basidiospores broadly ellipsoid to ellipsoid, (2.5–)2.9–4.2 × 2.0–3.2(–3.4) μm, L = 3.3 μm, W = 2.6 μm, Q = 1.08–1.60, smooth, hyaline, thin-walled, a few bearing droplets. Cystidia absent. Hyphal system monomitic, generative hyphae septate, branched, with clamp connections, thick-walled, hyaline, 2.6–4.8 μm in diam.
Specimens examined: Korea. Jeollanam-do, Gwangju Metropolitan City, Mudeungsan National Park, 35°06′21″N, 126°59′53″E, 14 Jul 2021, M. Cho, S. L. Kwon & Y. Yoo, KUC20210714-74 (=NIBRFG0000510827); Korea. Jeollanam-do, Gwangju Metropolitan City, Mudeungsan National Park, 35°15′26″N, 126°99′04″E, 17 Mar 2023, M. Cho, S. L. Kwon, S. H. Lee & D. Y. Kwon, KUC2023031701 (=NIBRFG0000514708).
Habitat: Occurs on dead trees in mixed hardwood forest.
Notes: Anomoloma luteoalbum is characterized by angular pores, abundant yellow rhizomorphs, absence of cystidia, and ellipsoid basidiospores. The basidiome of KUC20210714-14 appeared more yellowish, with thicker walls of basidia and basidioles compared to the type specimen (Cui 2687) [36]. The resupinate form of A. luteoalbum resembles A. rhizosum, though the latter has larger basidiospores (4.1–5.3 × 3–4 μm) [37]. In the phylogenetic tree, A. flavissimum is closely related to A. luteoalbum. Both species share features, such as yellow rhizomorphs and bright chrome or sulfur-yellow pore surfaces [38]. However, A. flavissimum can be distinguished by its larger basidiospores (3–4 × 2–3 μm) and the presence of cystidia [38]. Initially published as A. luteoalba [36], it was later revised to A. luteoalbum [39].
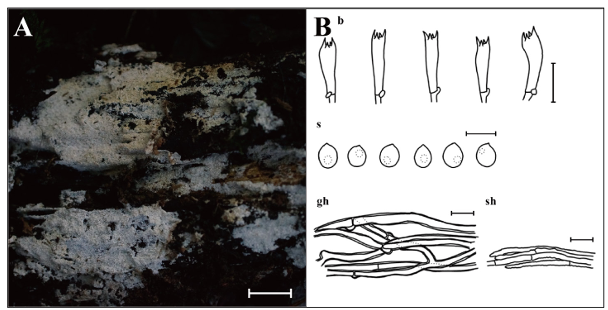
Fig. 6. Morphological characteristics of Fibrodontia alba KUC20210930-17. A: Basidiome; B: Microscopic structures, b: basidia; gh: generative hyphae; s: basidiospores; sh: skeletal hyphae (Scale bars: A = 1 cm, B = 10 μm, 5 μm for s).
MycoBank number: MB 564320
Basidiome effused, odontoid, soft when fresh, fragile when dry, covered with abundant aculei, 6–10 per mm, up to 0.4 mm long. Hymenial surface white (7.5YR, 9.5/1) when fresh, pale yellow (2.5Y, 8.5/2) when dry. Margin fimbriate or abrupt. Basidia with 4–spored, cylindrical to subclavate, 7.6–13.4 × 2.3–4.6μm, smooth, hyaline, with a basal clamp connection. Basidiospores subglobose to ellipsoid, (2.8–)2.9–3.9 × 2.2–3.3 μm, L = 3.3 μm, W = 2.7 μm, Q = 1.03–1.38, smooth, hyaline, thin-walled, bearing a droplet, with acute apex. Cystidia absent. Hyphal system pseudodimitic, generative hyphae septate, branched, with clamp connections, thick-walled, hyaline, 2.3–3.1 μm in diam. Skeletal-like hyphae septate, unbranched, thin- or thick-walled up to 1 μm with round apex, encrusted, hyaline, 1.7–2.6 μm in diam.
Specimen examined: Korea. Jeollanam-do, Gwangju Metropolitan City, Mudeungsan National Park, 35°05′90″N, 126°59′28″E, 30 Sep 2021, M. Cho, S. L. Kwon & Y. Yoo, KUC20210930-17 (=NIBRFG0000518401).
Habitat: Occurs on conifer tree stump in mixed hardwood forest.
Notes: Fibrodontia alba is characterized by an odontoid basidiome covered with numerous slender aculei and skeletal-like hyphae encrusted with crystals. The morphology of our specimen closely resembles the type specimen (TNM F24944), though observed basidia were shorter than those in the original description (11–19 × 3–3.5(–4.5) μm) [28]. Phylogenetic analysis indicates that F. subalba is most closely related to F. alba. Fibrodontia subalba shares an odontoid hymenophore and a white hymenial surface with F. alba [40] but differs in having a dimitic hyphal system and larger basidiospores ((3.5–)3.7–4.4(–4.6) × (2.7–)2.83.4(–3.5) µm) [40]. Interestingly, although F. alba was found on a conifer stump in this survey, it was reported on broad-leaved trees in the original description [28]. This species was previously reported only in central Taiwan [28]. This is the first report of F. alba in a temperate region and the first record of the genus Fibrodontia in this country.
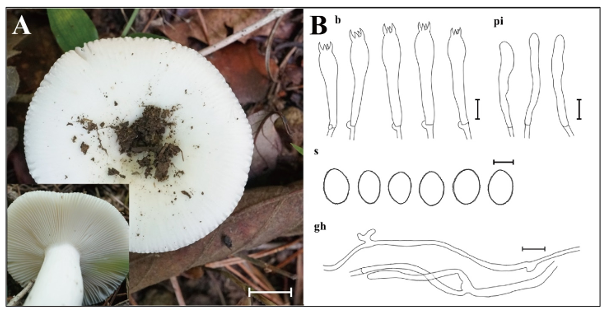
Fig. 7. Morphological characteristics of Russula pallidula KUC20220715-36. A: Basidiome; B: Microscopic structures, b: basidia; gh: generative hyphae; pi: pileocystidia; s: basidiospores (Scale bars: A = 1 cm, B = 10 μm, 5 μm for s).
MycoBank number: MB 825075
Pileus smooth, convex to plane, center depressed, white (10YR, 9.5/1), 40–50 mm in diam. Lamellae adnate, subfree, subcrowded to crowded, white (10YR, 9.5/1). Stipe central, equal, smooth, concolorous with pileus, 45 × 12 mm. Basidia with 4–spored, clavate to subclavate, 47.1–53.4 × 9.7–11.9 μm, smooth, hyaline, thin-walled. Basidiospores broadly ellipsoid to ellipsoid, asperulate, 7.5–9.8 × 5.8–7.3 μm, L = 8.7 μm, W = 6.5 μm, Q = 1.18–1.50, olive brown (2.5Y, 4/3). Pileocystidia narrowly cylindrical, 26.2–41.2 × 4.2–7.1 μm, smooth, hyaline, thin-walled. Hyphal system monomitic, generative hyphae septate, branched, thin-walled, hyaline, 2.5–3.5 μm in diam.
Specimen examined: Korea. Jeollanam-do, Hwasun-gun, Mudeungsan National Park, 35°10′29″N, 126° 98′38″E, 15 Jul 2022, M. Cho, S. L. Kwon & S. H. Lee, KUC20220715-36 (=NIBRFG0000518398).
Habitat: Solitary in the mixed hardwood forest.
Notes: Russula pallidula is characterized by white pileus and ellipsoid, asperculate basidiospores. It is classified under Russula subsect. Virescentinae. The morphological features of KUC20220715-36 generally align with those of the type specimen (RITF2613) [29], though cheilocystidia and pleurocystidia were rarely observed in the collected specimen. Morphologically and phylogenetically, R. sribuabanensis is closely related to R. pallidula. However, R. sribuabanensis is distinguishable by its larger pileus (2.5–10.5 cm) and subglobose basidiospores (6.2–8.2 × 5.6–7.2 μm, Q = 1.0–1.2) [41]. Previously reported from Southern China (Guang dong, Yunnan, and Zhejiang) [29], and this is the first report of R. pallidula in the temperate region.
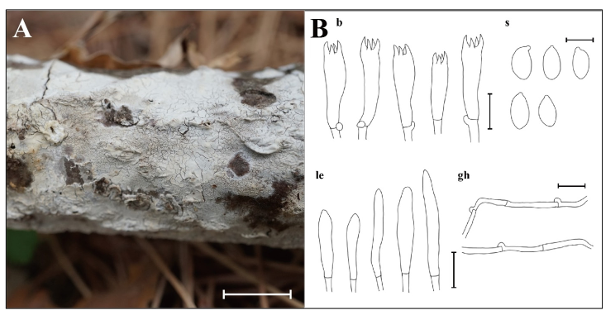
Fig. 8. Morphological characteristics of Schizocorticium parvisporum KUC20220414-13 and KUC20220414-22. A: Basidiome; B: Microscopic structures, b: basidia; gh: generative hyphae; le: leptocystidia; s: basidiospores (Scale bars: A = 1 cm, B = 10 μm, 5 μm for s).
MycoBank number: MB 830193
Basidiome adnate, widely effused, resupinate, membranous, with cracks. Hymenial surface smooth or sometimes irregular, white (7.5YR, 9.5/1) when fresh, pale yellow (2.5Y, 8.5/2) when dried. Hyphal system monomitic, generative hyphae septate, with clamp connections, thin-walled, hyaline, 2.1–3.3 μm in diam. Basidia with 4–spored, subclavate to cylindrical, 16.2–25.0 × 5.2–7.0 μm, smooth, hyaline, thin-walled, with a basal clamp connection. Basidiospores narrowly ellipsoid to cylindrical, 6.1–6.8 × 3.1–4.0 μm, L = 6.3 μm, W = 3.6 μm, Q = 1.6–1.9, smooth, hyaline, thin-walled, with acute apex. Cystidia absent. Hyphidia tubular, narrowly cylindrical, smooth, thin-walled, hyaline, 15.8–32.9 × 2.2–3.9 μm.
Specimens examined: Korea. Jeollanam-do, Hwasun-gun, Mudeungsan National Park, 14 Apr 2022, 35° 10′35″N, 126°98′81″E, M. Cho, S. L. Kwon & S. H. Lee, KUC20220414-13 (=NIBRFG0000513227); Korea. Jeollanam-do, Hwasun-gun, Mudeungsan National Park, 35°10′33″N, 126°98′79″E, 14 Apr 2022, M. Cho, S. L. Kwon & S. H. Lee, KUC20220414-22 (=NIBRFG0000513233).
Habitat: Occurs on the dead angiosperm branches in the hardwood forest.
Notes: Schizocorticium parvisporum is characterized by effused and cracked basidiome, and the presence of hyphidia. It is typically found in angiosperm branches or trunks [27]. The studied specimens share a similar morphology with the type specimen (GC 1508-127), although numerous branched hyphidia were observed in the type specimen [27]. Morphological similarities are common among Schizocorticium species [27], but they can be differentiated by basidiospore size: S. magnosporum has the largest basidiospores (9.8–11.5 × 4.2–5.5 μm), followed by S. mediosporum (8–9.8 × 3.8–4.8 μm), and S. parvisporum with the smallest [27]. Previous research has emphasized the necessity of tef1 region analysis for precise identification of Schizocorticium species [27]. For instance, in the phylogenetic tree, S. mediosporum is the closest to S. parvisporum, with only 18 bp differences in the ITS and nLSU combined dataset. This limited variation caused an unclear separation when using these two regions alone. However, in the tef1 region, the difference increased significantly to 70 bp, thereby facilitating better resolution. Therefore, combined phylogenetic analyses using the ITS+nLSU+tef1 datasets are essential for the accurate identification of Schizocorticium species. Previously known from subtropical Taiwan and Yunnan, China [27], this is the first report of S. parvisporum in the temperate region.
The authors declare that this study was conducted in the absence of any commercial or financial relationships that could be construed as potential conflicts of interest.
This research was supported by the National Institute of Biological Resources [NIBR202402104] under the Ministry of the Environment, Republic of Korea. This study was also supported by the Korea University Grant.
1. Van Geffen KG, Poorter L, Sass-Klaassen U, Van Logtestijn RS, Cornelissen JH. The trait contribution to wood decomposition rates of 15 Neotropical tree species. Ecology 2010;91:3686-97. [DOI]
2. Bani A, Pioli S, Ventura M, Panzacchi P, Borruso L, Tognetti R, Tonon G, Brusetti L. The role of microbial community in the decomposition of leaf litter and deadwood. Appl Soil Ecol 2018;126:75-84. [DOI]
3. Smith SE, Read DJ. Mycorrhizal symbiosis. Cambridge: Academic press; 2010.
4. Policelli N, Horton TR, Hudon AT, Patterson TR, Bhatnagar JM. Back to roots: the role of ectomycorrhizal fungi in boreal and temperate forest restoration. Front For Glob Change 2020;3:97. [DOI]
5. Bilal AW, Bodha RH, Wani AH. Nutritional and medicinal importance of mushrooms. J Med Plants Res 2010;4:2598-604. [DOI]
6. Wu B, Hussain M, Zhang W, Stadler M, Liu X, Xiang M. Current insights into fungal species diversity and perspective on naming the environmental DNA sequences of fungi. Mycology 2019;10:127-40. [DOI]
7. Sridhar KR, Deshmukh SK. Advances in macrofungi: diversity, ecology and biotechnology. London: CRC Press; 2019. 8. Hawksworth DL, Lücking R. Fungal diversity revisited: 2.2 to 3.
8 million species. Microbiol Spectr 2017;5:10.1128/microbiolspec.funk-0052-2016.
9. Hawksworth DL. The magnitude of fungal diversity: the 1.5 million species estimate revisited. Mycol Res 2001;105:1422-32. [DOI]
10. National Institute of Biological Resources (NIBR). National list of species of Korea [Internet]. Incheon: NIBR; 2023 [cited 2024 Dec 5]. Available from http://kbr.go.kr.
11. Han S, Cho Y, Shin Y, Seo S, Lee D. Avifauna of the Mudeungsan (Mt.) region of Gwangju Metropolitan City. J Korean Nat 2011;4:35-43. [DOI]
12. Ko MH, Jang SL, Won YJ. Fish distribution characteristics of Mudeungsan National Park. Korean J. Environ Ecol 2018;32:154-64. [DOI]
13. Lee JW, Lee S, Kang SH. A study on the flora and its introduced disturbing plants in Damyang area of Mudeungsan National Park, Korea. Korean J Plant Res 2021;34:103-13.
14. Jeong J, Han S, Kwon Y. Plant sociological relationships around the bedrock to conserve sustainable vegetation area at the summit of Mudeungsan National Park in South Korea. Environ Dev Sustain 2023;25:7851-71. [DOI]
15. Cho M, Kwon SL, Heo YM, Lee YM, Lee H, Kim C, Ahn BJ, Kim JJ. Seven unrecorded indigenous fungi from Mudeungsan National Park in Korea. Mycobiology 2022;50:203-12. [DOI]
16. Gardes M, Bruns TD. ITS primers with enhanced specificity for basidiomycetes‐application to the identification of mycorrhizae and rusts. Mol Ecol 1993;2:113-8. [DOI]
17. White TJ, Bruns TD, Lee SB, Taylor JW. Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics. In: Innis MA, Gelfand DH, Sninsky JJ, editors. PCR protocols: A guide to methods and applications. San Diego: Academic Press; 1990. p. 315-22. [DOI]
18. Vilgalys R, Hester M. Rapid genetic identification and mapping of enzymatically amplified ribosomal DNA from several Cryptococcus species. J Bacteriol 1990;172:4238-46. [DOI]
19. Hopple Jr JS, Vilgalys R. Phylogenetic relationships among coprinoid taxa and allies based on data from restriction site mapping of nuclear rDNA. Mycologia 1994;86:96-107. [DOI]
20. Rehner SA, Buckley E. A Beauveria phylogeny inferred from nuclear ITS and EF1-α sequences: evidence for cryptic diversification and links to Cordyceps teleomorphs. Mycologia 2005;97:84-98. [DOI]
21. Sayers EW, Cavanaugh M, Clark K, Pruitt KD, Sherry ST, Yankie L, Karsch-Mizrachi I. GenBank 2024 update. Nucleic Acids Res 2024;52(D1):D134-D7. [DOI]
22. Katoh K, Standley DM. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol Evol 2013;30:772-80. [DOI]
23. Miller MA, Schwartz T, Pickett BE, He S, Klem EB, Scheuermann RH, Passarotti M, Kaufman S, O’Leary MA. A RESTful API for access to phylogenetic tools via the CIPRES Science Gateway. Evol Bioinform Online 2015;11:43-8. [DOI]
24. Stamatakis A. RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics 2014;30:1312-3. [DOI]
25. Rambaut A. FigTree, a graphical viewer of phylogenetic trees (Version 1.4.4). Edinburgh: Institute of Evolutionary Biology, University of Edinburgh; 2018.
26. X-Rite. Munsell soil-color charts with genuine Munsell® color chips. Grand Rapids: X-Rite; 2009.
27. Wu SH, Wei CL, Chen YP, Chen CC, Chen SZ. Schizocorticium gen. nov. (Hymenochaetales, Basidiomycota) with three new species. Mycol Prog 2021;20:769-79. [DOI]
28. Yurchenko E, Wu SH. Fibrodontia alba sp. nov. (Basidiomycota) from Taiwan. Mycoscience 2014;55:336-343. [DOI]
29. Chen B, Jiang X, Song J, Liang J, Wang S, Lu J. Morphological and phylogenetic analyses reveal the new species Russula pallidula from China. Sydowia 2019;71:1-10.
30. Jaroszewicz B, Cholewińska O, Chećko E, Wrzosek M. Predictors of diversity of deadwooddwelling macrofungi in a European natural forest. For Ecol Manag 2021;490:119123. [DOI]
31. Schmit JP, Mueller GM, Leacock PR, Mata JL, Wu Q, Huang Y. Assessment of tree species richness as a surrogate for macrofungal species richness. Biol Conserv 2005;121:99-110. [DOI]
32. Zhang Y, Zhou DQ, Zhao Q, Zhou TX, Hyde KD. Diversity and ecological distribution of macrofungi in the Laojun Mountain region, Southwestern China. Biodivers Conserv 2010;19:3545-63. [DOI]
33. Yu H, Wang T, Skidmore A, Heurich M, Bässler C. How future climate and tree distribution changes shape the biodiversity of macrofungi across Europe. Divers Distrib 2023;29:666-82. [DOI]
34. Kim CC, Kim TG. Evaluation on climate change vulnerability of Korea National Parks. Korea J Ecology Environ 2016;49:42-50. [DOI]
35. Adhikari P, Shin MS, Jeon JY, Kim HW, Hong S, Seo C. Potential impact of climate change on the species richness of subalpine plant species in the mountain national parks of South Korea J Ecology Environ 2018;42:36. [DOI]
36. Song J, Liu XY, Wang M, Cui BK. Phylogeny and taxonomy of the genus Anomoloma (Amylocorticiales, Basidiomycota). Mycol Prog 2016;15:1-8. [DOI]
37. Niemelä T, Larsson KH, Dai YC, Larsson E. Anomoloma, a new genus separated from Anomoporia on the basis of decay type and nuclear rDNA sequence data. Mycotaxon 2007;100:305-17.
38. Ryvarden L. East Asian Polypores. Volume 2, Polyporaceae s.l. Oslo: Fungiflora; 2001.
39. Zhou M, Vlasák J, Ghobad-Nejhad M, Lim YW, Dai YC. Taxonomy and an updated phylogeny of Anomoloma (Amylocorticiales, Basidiomycota). Forests 2022;13:713. [DOI]
40. Liu SL, He SH, Liu DM, Zhou LW. Two new species of Fibrodontia (Trechisporales, Basidiomycota) with a key to worldwide species. J Fungi 2021;7:982. [DOI]
41. Paloi S, Kumla J, Karunarathna SC, Lumyong S, Suwannarach N. Taxonomic and phylogenetic evidence reveal two new Russula species (Russulaceae, Russulales) from Northern Thailand. Mycol Prog 2023;22:72. [DOI]