Jun-Woo Choi1, Gwang-Jae Lim1, Chang-Gi Back2, In-Kyu Kang3, Seung-Yeol Lee1,4*, and Hee-Young Jung1,4
1Department of Plant Medicine, Kyungpook National University, Daegu 41566, Korea
2Department of Environmental Horticulture and Landscape Architecture, Environmental Horticulture, Dankook University, Cheonan 31116, Korea
3Department of Horticultural Science, Kyungpook National University, Daegu 41566, Korea
4Institute of Plant Medicine, Kyungpook National University, Daegu 41566, Korea
*Correspondence to leesy1123@knu.ac.kr
Korean Journal of Mycology (Kor J Mycol) 2025 March, Volume 53, Issue 1, pages 1-9.
https://doi.org/10.4489/kjm.2025.53.1.1
Received on January 31, 2025, Revised on February 17, 2025, Accepted on February 24, 2025, Published on Mar 31, 2025.
Copyright © The Korean Society of Mycology.
This is an Open Access article which is freely available under the Creative Commons Attribution-NonCommercial 4.0 International License (CC BY-NC) (https://creativecommons.org/licenses/by-nc/4.0/).
A fungal strain was isolated from a darkened apple tree trunk and designated as KNUF21-C5. The cultural and morphological characteristics of the isolated strain were evaluated using potato dextrose and malt extract agar (MEA). After 7 days, the colonies on MEA were circular, dense, rough, lacking aerial mycelia, olivaceous-yellow at the center with white margins, and reached 90 mm in diameter. Morphologically, the conidiophores were straight and hyaline, and the conidiogenous cells were enteroblastic, phialidic, hyaline, and cylindrical. The conidia were unicellular, elongated-allantoid, and measured 5.1–6.6 × 1.3–1.6 µm in diameter. These characteristics were consistent with those of the Cytospora species. To identify the species, molecular analysis was performed using sequences of the internal transcribed spacer regions, large subunit of 28S rRNA, actin, translation elongation factor 1-alpha, and RNA polymerase II subunit gene, it showed 98.0–100% similarity to C. erumpens CFCC 53163. The KNUF-21-C5 strain was clustered with C. erumpens CFCC 53163 in phylogenetic trees, and the conidial size was similar to the type strain of C. erumpens (5.1–6.6 × 1.3–1.6 µm vs. 5.6–6.7 × 1.3–1.7 µm). Based on fungal characteristics and phylogenetic analysis, KNUF-21-C5 was identified as C. erumpens. The pathogenicity of KNUF-21-C5 in apples was confirmed by inoculation of apple twigs. This is the first record of C. erumpens associated with Cytospora canker on apples in Korea.
Apple, Cytospora canker, Cytospora erumpens, Morphology, Phylogenetic analysis
Apple (Malus × domestica Borkh.) is one of the most economically significant fruits cultivated all around the world [1]. In Korea, apples are highly valuable fruits grown on 33,313 ha, with a production of 460,088 tons in 2024 [2]. The production and quality of apples can be affected by various fungal pathogens, and some species, including Diplodia, Neofusicoccum, Botryosphaeria, and Cytospora, cause canker and dieback symptoms in apple trees and twigs [3,4]. Among these, Cytospora canker (also called Valsa canker), caused by Cytospora species, was first reported on apples in Japan in 1903 and is one of the most destructive diseases that cause significant economic losses in East Asia [5]. The genus Cytospora (Diaporthales) was established by Ehrenberg in 1818, and more than 100 species have been described as phytopathogens, endophytes, and saprobes [6]. Cytospora is associated with several sexual genera such as Valsa, Leucocytospora, Lecuostoma, and Valsella [7]. In 2005, these sexual genera were synonymized under Valsa by Adams et al., and they were classified as the oldest name, Cytospora following the application of the “one fungus-one name” principle [7]. Cytospora canker disease can occur when the host is weakened by abiotic stress, such as during droughts, by colonizing the bark of host trees through wounds and other mechanical injuries, and extensively invading into phloem and xylem [6,8]. The canker begins with water-soaked symptoms on trunks or branches, and the epidermis turns reddish brown [9]. The lesions may later dry, forming sunken, dark brown cankers that can cause the death of the entire tree. Fruiting bodies are formed on infected hosts and yellow-orange to red spore tendrils can exude from fruiting bodies under moist conditions [7]. The fruiting bodies contained single or labyrinthine locules with hyaline conidiophores and allantoid conidia. During an apple disease survey in 2021, a sample of a declined apple tree was collected from an apple orchard in Gyeonggi Province, Korea. In this study, a Cytospora species was isolated from apple trunks and identified based on its cultural and morphological characteristics, along with a phylogenetic analysis.
Apple tree samples (cv. Gamhong) showing decline and darkening symptoms were collected from an apple orchard in Paju-si, Gyeonggi Province, Korea. To isolate the causal agent, the bark from the darkened trunk was removed using a scalpel. Then, the pieces of dark-brown tissue on the trunk were transferred onto potato dextrose agar (PDA; Difco, Detroit, MI, USA), and incubated at 25℃ in the dark. After a few days, the margins of the colonies were sub-cultured on fresh PDA. The isolated strain was designated as KNUF-21-C5 and was maintained in 20% glycerol at –80℃ for further study. The isolated strain has been deposited in the Korean Agricultural Culture Collection (KACC410961).
The strain KNUF-21-C5 was cultured on PDA and malt extract agar (MEA; Difco, Detroit, MI, USA) for 7 days at 25℃ to observe the cultural and morphological characteristics. Mycelial plugs were extracted from colonies grown on PDA using a 4 mm cork borer. The conidiophores, conidiogenous cells, and conidia in the pycnidia were observed and measured using an optical microscope (BX-50; Olympus, Tokyo, Japan). Fungal growth was quantified by measuring the diameter of colonies using Vernier calipers (Mitutoyo, Kawasaki, Japan).
Total genomic DNA was extracted from the strain grown on PDA using the HiGene™ Genomic DNA Prep Kit (BIOFACT, Daejeon, Korea) following the manufacturer’s protocol. The internal transcribed spacer (ITS) regions, large subunit of 28S rRNA (LSU), actin (ACT), translation elongation factor 1-alpha (TEF1), and RNA polymerase II subunit (RPB2) genes of the strain were amplified using primer pairs of ITS1F/ITS4, LROR/LR7, ACT-512F/ACT-783R, EF1-688F/EF1-1251R, and RPB2-5F2/fRPB2-7cR, respectively [10–17]. The PCR products were analyzed by gel electrophoresis and stained with ethidium bromide. The purification of amplified products was conducted using ExoSAP-IT (Thermo Fisher Scientific, Waltham, MA, USA) and sequenced by Solgent (Daejeon, Korea). The obtained sequences of the ITS regions, LSU, ACT, TEF1, and RPB2 were submitted to the National Center for Biotechnology Information (NCBI) (Table 1).
Table 1. GenBank accession numbers of Cytospora species used for phylogenetic analysis
| Species | Strain number | Isolation source | Origin | GenBank Accession number | ||||
|---|---|---|---|---|---|---|---|---|
| ITS | LSU | ACT | TEF1 | RPB2 | ||||
| Cytospora albodisca | CFCC 53161T | Platycladus orientalis | China | MW418406 | MW418418 | MW422899 | MW422921 | MW422909 |
| C. albodisca | CFCC 54373 | Platycladus orientalis | China | MW418407 | MW418419 | MW422900 | MW422922 | MW422910 |
| C. ceratospermopsis | CFCC 89626T | Juglans regia | China | KR045647 | KR045726 | KU711011 | KU710934 | KU710978 |
| C. ceratospermopsis | CFCC 89627 | Juglans regia | China | KR045648 | KR045727 | KU711012 | KU710935 | KU710979 |
| C. erumpens | CFCC 53163 | Prunus padus | China | MK673059 | MK673089 | MK673029 | MK672948 | MK673000 |
| C. erumpens | KNUF-21-C5 | Malus domestica | Korea | PQ821439 | PQ821445 | PQ827002 | PQ827004 | PQ827003 |
| C. gigaspora | CFCC 50014 | Juniperus procumbens | China | KR045630 | KR045710 | KU710999 | KU710922 | KU710959 |
| C. gigaspora | CFCC 89634T | Salix psammophila | China | KF765671 | KF765687 | KU711000 | KU710923 | KU710960 |
| C. leucostoma | CFCC 53140 | Prunus sibirica | China | MN854445 | MN854656 | MN850760 | MN850753 | MN850746 |
| C. leucostoma | CFCC 53141 | Prunus sibirica | China | MN854446 | MN854657 | MN850761 | MN850754 | MN850747 |
| C. mali | ARI-15-US | Malus domestica | Korea | PP974558 | PP976972 | LC830507 | LC830505 | LC830509 |
| C. mali | ARI-23-GW | Malus domestica | Korea | PP974557 | PP976971 | LC830506 | LC830504 | LC830508 |
| C. mali | CFCC 50030 | Malus pumila | China | MH933643 | MH933677 | MH933550 | MH933524 | MH933608 |
| C. mali | CFCC 50031 | Crataegus sp. | China | KR045636 | KR045716 | KU711004 | KU710927 | KU710965 |
| C. mali-spectabilis | CFCC 53181T | Malus spectabilis | China | MK673066 | MK673096 | MK673036 | MK672953 | MK673006 |
| C. nivea | CFCC 89641 | Elaeagnus angustifolia | China | KF765683 | KF765699 | KU711006 | KU710929 | KU710967 |
| C. olivacea | CFCC 53176T/ | Sorbus tianschanica | China | MK673068 | MK673098 | MK673038 | MK672955 | MK673008 |
| Diaporthe eres | CBS 145040 | Lactuca satia | Netherlands | MK442579 | MK442521 | MK442634 | MK442693 | MK442663 |
ITS: internal transcribed spacer regions; LSU: large subunit of 28S rRNA; ACT: actin; TEF1: translation elongation factor 1-alpha; RPB2: RNA polymerase II subunit.
T type strain.
The strain identified in this study is indicated in bold.
Phylogenetic analysis was performed using sequences obtained from the NCBI database (Table 1). The ambiguous regions were removed from the alignments, and the evolutionary distance matrices were calculated using the maximum-likelihood (ML) method with ClustalX and the Tamura-Nei model [18]. Phylogenetic trees were constructed using the ML method in MEGA 11.0, with bootstrap values derived from 1,000 replications [19].
To test Koch’s postulates, a pathogenicity test was conducted by inoculating apple twigs with the KNUF21-C5 strain according to the inoculation method described previously [7]. The twigs were washed with tap water, sterilized with 70% ethanol for 4 min, and dried for 2 min. The bark of the twigs was removed using a disinfected scalpel and inoculated with a 4 mm diameter mycelial plug from a 5-day-old colony grown on PDA. The inoculated twigs were incubated in a moist chamber at 25℃ for 2 weeks. After incubation, small pieces from the inoculation sites were transferred onto PDA medium to re-isolate the inoculated strain.
The colonies of the KNUF-21-C5 strain on PDA were flat, circular, irregular, lacking aerial mycelia, yellow to cream at the center, white toward the edges, and reached 90 mm after 3 days. On MEA, the colonies exhibited a circular form, dense mycelia, and a rough, irregular, subhyaline texture with white margins. The center of the colonies was yellow to olivaceous-yellow, reaching a diameter of 90 mm after 7 days. Colonies were flat and lacked aerial mycelia. The reverse side of the colonies also displayed bright khaki at the center and white margins (Fig. 1A–D). Pycnidia, exudating dark orange to brownish masses of conidia, were observed in the colonies (Fig. 1E). Conidiophores were straight, cylindrical, unbranched, or occasionally branched at the base and were reduced to conidiogenous cells. Conidiogenous cells formed within the pycnidial wall, appearing hyaline, blastic, enteroblastic, phialidic, and cylindrical with smooth walls (Fig. 1F). Conidia were unicellular, hyaline, allantoid to subcylindrical, slightly curved, and smooth-walled, and typically measured 5.1–6.6 × 1.3–1.6 µm (avg. 5.6 × 1.4 µm, n = 50) (Fig. 1G). The culture and morphological characteristics of KNUF-21-C5 were similar to those previously reported for C. erumpens [20].
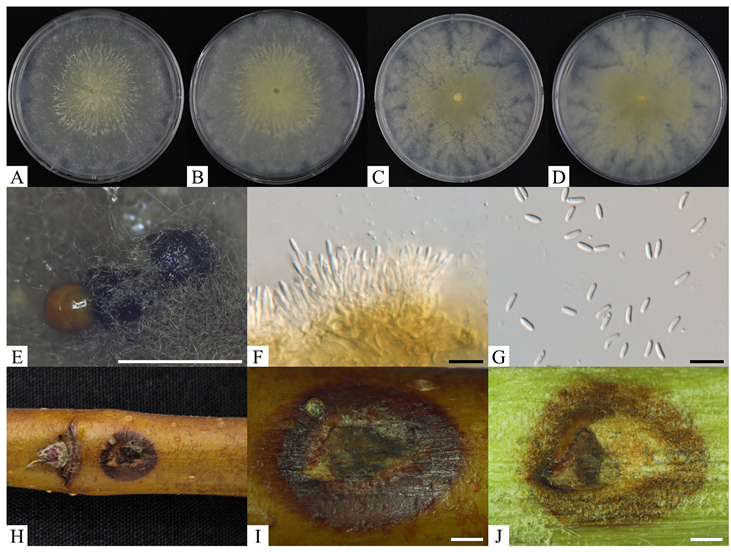
Fig. 1. Morphological characteristics of Cytospora erumpens KNUF-21-C5 isolated from apple trunk. A, B: obverse and reverse of colony on PDA at 25℃ for 7 days, respectively; C, D: obverse and reverse of colony on MEA at 25℃ for 7 days, respectively; E: pycnidia exudating conidial mass; F: conidiophores with conidiogenous cells; G: conidia; H–J: result of pathogenicity test for 2 weeks. White scale bars = 1 mm, black scale bars = 10 μm. PDA: potato dextrose agar; MEA: malt extract agar.
Partial sequences of the ITS regions, LSU, ACT, TEF1, and RPB2 were amplified, yielding sequences of 616, 1,262, 248, and 981 bp, respectively. The ITS sequence showed 99.8% similarity with C. erumpens CFCC 53163 and C. leucostoma CFCC 54140. The LSU sequence exhibited 100% similarity with C. erumpens CFCC 53163 and 99.4% similarity with C. leucostoma 54140 and C. albodisca CFCC 53161T. The partial ACT sequence showed 98.4% similarity with C. erumpens CFCC 53163 and less than 93.2% similarity with other Cytospora species, including C. leucostoma CFCC 54140 and C. albodsica CFCC 53161T. In the case of TEF1, 98.0% similarity with C. erumpens CFCC 53163 and 88.2% similarity with C. leucostoma CFCC 53140. The RPB2 sequence revealed 98.5% similarity with C. erumpens CFCC 53163, and 94.6–95.9% similarity with C. leucostoma and C. albodisca strains. The ML phylogenetic tree was constructed based on the concatenated sequences of the ITS regions, LSU, ACT, TEF1, and RPB2. The KNUF-21-C5 strain clustered with C. erumpens CFCC 53163, forming a cluster distinct from other Cytospora species in the phylogenetic tree (Fig. 2).
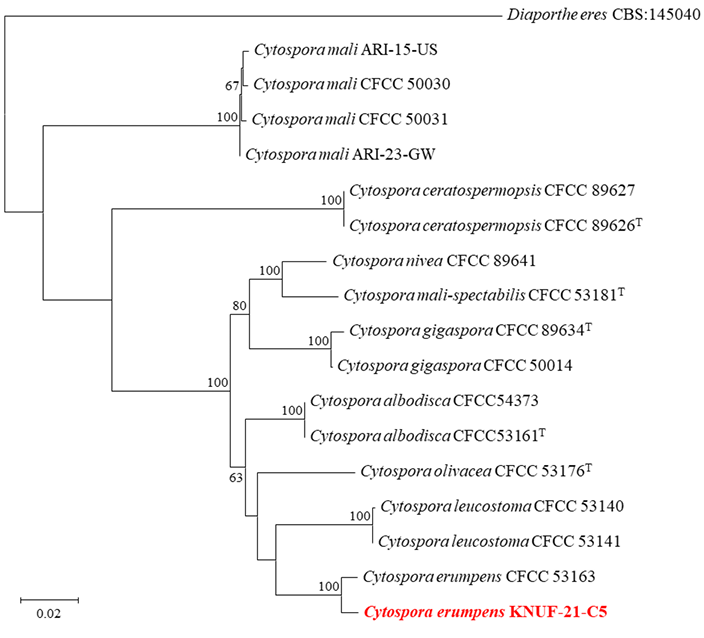
Fig. 2. Maximum-likelihood phylogenetic tree based on concatenated sequences of ITS regions, LSU, ACT, TEF1, and RPB2, showing the relationship between the strain KNUF-21-C5 with closest Cytospora species. The numbers above the branches indicate bootstrap values (> 60%) obtained from 1,000 replicates. Diaporthe eres CBS 145050 was used as an outgroup. The isolated strain in this study is indicated in red and bold. Bar = 0.02 substitutions per nucleotide position. T type strain. ITS: internal transcribed spacer regions; LSU: large subunit of 28S rRNA; ACT: actin; TEF1: translation elongation factor 1-alpha; RPB2: RNA polymerase II subunit.
Discoloration and browning symptoms were observed on the bark at the inoculation sites two weeks post-inoculation (Fig. 1H–J). A fungal strain was re-isolated from lesions at the inoculation site, and its cultural and morphological characteristics were identical to those of the strain used for inoculation. In addition, the partial sequence of the re-isolated strain matched that of KNUF-21-C5 (data not shown). The pathogenicity of KNUF-21-C5 was confirmed on apple twig under laboratory conditions, though it showed weak pathogenicity.
The genus Cytospora includes several plant pathogens that cause Cytospora canker diseases, primarily affecting woody hosts with a wide distribution and broad host range [9]. Traditionally, the identification of Cytospora species that cause Cytospora canker has been based on morphological characteristics and host affiliation [20]. However, recent studies have shown that several Cytospora species such as C. chrysosperma and C. punicae infect a range of hosts rather than being host-specific [21]. C. erumpens was f irst described as causing cankers and dieback on twigs and branches of crack willows (Salix fragilis) in Russia in 2017 [20]. Subsequently, C. erumpens was isolated from canker lesions on bird cherries (Prunus padus), peaches (P. persica), and apples (Malus spp.) in China [7,22,23]. The pathogenicity of Cytospora species on apple varieties and wild apples was evaluated, including C. erumpens [23]. The results showed that the pathogenicity of Cytospora species varied depending on the variety, and C. erumpens was strongly pathogenic to apples in the leaf test, with relatively weak pathogenicity to branches [23]. According to Lin et al., C. erumpens is regarded as a synonym of C. leucostoma [24]. However, recent research has shown that C. erumpens forms a cluster distinct from C. leucostoma strains in phylogenetic trees [23]. Moreover, the strain of C. erumpens was differentiated from C. leucostoma CFCC 53140 based on cultural and morphological characteristics, such as colony color, ostiole diameter, and conidial size [23]. In this study, the strain KNUF-21-C5 exhibited distinctive cultural characteristics compared to C. leucostoma CFCC 53140 (Table 2). Additionally, the strain clustered with C. erumpens CFCC 53163, but was distinct from C. leucostoma CFCC 53140 (Fig. 2). Therefore, C. erumpens KNUF-21-C5 differed from C. leucostoma.
Cytospora canker is one of the most significant diseases in apple trees, and over 20 Cytospora species have been reported in apples, including C. chrysosperma, C. leucostoma, C. nivea, and C. pyri [6,25]. According to a survey conducted from 2015 to 2024, the incidence of apple Cytospora canker has sharply increased during the last three years, and it has been confirmed that C. mali is predominant in apple orchards in Korea [25]. Recently, the distribution of fungi has been affected by global warming, and the distributions of C. chrysosperma, C. mali, and C. nivea are expected to shift toward the northeastern regions of China [26]. For this reason, other Cytospora species are presumed to inhabit apple orchards in Korea, although C. mali is predominant. In this study, KNUF-21-C5 was isolated from the trunks of apple trees in Korea. Based on its cultural and morphological characteristics, and phylogenetic analysis, the strain KNUF-21-C5 was identified as a previously undescribed species, C. erumpens. To our knowledge, this is the first report of C. erumpens isolated from apple trunks in Korea.
Table 2. Cultural and morphological characteristics of the isolated strain KNUF-21-C5 with reference to Cytospora species
| Characteristics | C. erumpens KNUF-21-C5a | C. erumpens MFLUCC 16-0580Tb | C. leucostoma CFCC 53140c | C. albodisca CFCC 53161Td | |
|---|---|---|---|---|---|
| Colony | PDA | Flat, lacking aerial mycelium, irregular, yellow, white margins, 90 mm at 3 days. | N/A | Flat, greenish-olivaceous to grey olivaceous, 90 mm at 4 days. | Sparse in the center, compact margin, felt, white to dark herbage green, 90 mm at 5 days. |
| MEA | Flat, lacking aerial mycelium, dense, olivaceous-yellow, white margins, 90 mm at 7 days. | Dense, circular, margin rough, white, lacking aerial mycelium, 85 mm at 7 days. | N/A | N/A | |
| Conidiophores | Straight, hyaline, unbranched at the base, or occasionally branched. | Unbranched or occasionally branched at the base. | Hyaline, branched at the base or unbranched, cylindrical. | Cylindrical, hyaline, unbranched, straight to slightly sinuous. | |
| Conidiogenous cells | Enteroblastic, phialidic, hyaline, cylindrical. | Enteroblastic, phialidic, hyaline. | Enteroblastic, phialidic, sub-cylindrical to cylindrical. | Enteroblastic, phialidic. | |
| Conidia | Unicellular, hyaline, elongated-allantoid, slightly curved, smooth, 5.1–6.6 × 1.3–1.6 µm. | Unicellular, elongate-allantoid, hyaline, smooth, 5.6–6.7 × 1.3–1.7 µm. | Hyaline, elongate-allantoid, smooth, aseptate, 4.5–6.0 × 1.0–2.0 µm. | Hyaline, allantoid, little curve, rough, aseptate, biguttulate, 5–7 × 1–2 μm. |
a fungal strain investigated in this study; b sources of description [20]; c sources of description [27]; d sources of description [21]; T type strain.
PDA: potato dextrose agar; MEA: malt extract agar; N/A: not available in previous study.
The authors declare that they have no potential conflicts of interest.
This research was supported by the “Cooperative Research Program for Agriculture Science and Technology Development” (Project No. RS-2024-00396930) of the Rural Development Administration, Republic of Korea.
1. Food and Agriculture Organization of the United Nations. Food and agriculture data [Internet]. Rome: Food and Agriculture Organization of the United Nations; 2023 [cited 2024 Dec 20]. Available from: https://www.fao.org/faostat/en/#data/QCL.
2. Korea Statistical Information Service. Fruits production [Internet]. Daejeon: Statistics Korea; 2024 [cited 2024 Dec 23]. Available from: https://kosis.kr.
3. Hanifeh S, Zafari D, Soleimani MJ, Arzanlou M. Multigene phylogeny, morphology, and pathogenicity trials reveal novel Cytospora species involved in perennial canker disease of apple trees in Iran. Fungal Biol 2022;126:707-26. [DOI]
4. Díaz GA, Valdez A, Halleen F, Ferrada E, Lolas M, Latorre BA. Characterization and pathogenicity of Diplodia, Lasiodiplodia, and Neofusicoccum species causing Botryosphaeria canker and dieback of apple trees in Central Chile. Plant Dis 2022;106:925-37. [DOI]
5. Wang X, Shi CM, Gleason ML, Huang L. Fungal species associated with apple Valsa canker in East Asia. Phytopathol Res 2020;2:35. [DOI]
6. Sha SS, Wang Z, Yan CC, Hao HT, Wang L, Feng HZ. Identification of fungal species associated with apple canker in Tarim Basin, China. Plant Dis 2023;107:1284-98. [DOI]
7. Fan XL, Bezerra JDP, Tian CM, Crous PW. Cytospora (Diaporthales) in China. Persoonia 2020;45:1-45. [DOI]
8. Azizi R, Ghosta Y, Ahmadpour A. Apple crown and collar canker and necrosis caused by Cytospora balanejica sp. nov. in Iran. Sci Rep 2024;14:6629. [DOI]
9. Petrović E, Vrandečić K, Ivić D, Ćosić J, Godena S. First report of olive branch dieback in Croatia caused by Cytospora pruinosa Défago. Microorganisms 2023;11:1679. [DOI]
10. Gardes M, Bruns TD. ITS primers with enhanced specificity for basidiomycetes-application to the identification of mycorrhizae and rusts. Mol Ecol 1993;2:113-8. [DOI]
11. White TJ, Bruns TD, Lee SB, Taylor JW. Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics. In: Innis MA, Gelfand DH, Sninsky JJ, editors. PCR protocols: A guide to methods and applications. San Diego: Academic Press; 1990. p. 315-22. [DOI]
12. Rehner SA, Samuels GJ. Taxonomy and phylogeny of Gliocladium analyzed from nuclear large subunit ribosomal DNA sequences. Mycol Res 1994;98:625-34. [DOI]
13. Vilgalys R, Hester M. Rapid genetic identification and mapping of enzymatically amplified ribosomal DNA from several Cryptococcus species. J Bacteriol 1990;172:4238-46. [DOI]
14. Carbone I, Kohn LM. A method for designing primer sets for speciation studies in filamentous ascomycetes. Mycologia 1999;91:553-6. [DOI]
15. Alves A, Crous PW, Correia A, Phillips AJL. Morphological and molecular data reveal cryptic speciation in Lasiodiplodia theobromae. Fungal Divers 2008;28:1-13.
16. Sung GH, Sung JM, Hywel-Jones NL, Spatafora JW. A multi-gene phylogeny of Clavicipitaceae (Ascomycota, Fungi): Identification of localized incongruence using a combinational bootstrap approach. Mol Phylogenet Evol 2007;44:1204-23. [DOI]
17. Liu YJ, Whelen S, Hall BD. Phylogenetic relationships among ascomycetes: evidence from an RNA polymerase II subunit. Mol Biol Evol 1999;16:1799-808. [DOI]
18. Tamura K, Nei M. Estimation of the number of nucleotide substitutions in the control region of mitochondrial DNA in humans and chimpanzees. Mol Biol Evol 1993;10:512-26.
19. Tamura K, Stecher G, Kumar S. MEGA11: Molecular evolutionary genetics analysis version 11. Mol Biol Evol 2021;38:3022-7. [DOI]
20. Norphanphoun C, Doilom M, Daranagama DA, Phookamsak R, Wen TC, Bulgakov TS, Hyde KD. Revisiting the genus Cytospora and allied species. Mycosphere 2017;8:51-97. [DOI]
21. Pan M, Zhu H, Tian C, Huang M, Fan X. Assessment of Cytospora isolates from conifer cankers in China, with the descriptions of four new Cytospora species. Front Plant Sci 2021;12:636460. [DOI]
22. Zhao Y, Cai G, Yan M, Ma R, Zhang D. Pathogenicity evaluation of Cytospora species in 13 apple (Malus domestica) varieties and wild apple (Malus sieversii) in Xinjiang, China. J Phytopathol 2024;172:e13375. [DOI]
23. He Z, Abeywickrama PD, Wu L, Zhou Y, Zhang W, Yan J, Shang Q, Zhou Y, Li S. Diversity of Cytospora species associated with trunk diseases of Prunus persica (peach) in Northern China. J Fungi 2024;10:843. [DOI]
24. Lin L, Fan XL, Groenewald JZ, Jami F, Wingfield MJ, Voglmayr H, Jaklitsch W, Castlebury LA, Tian CM, Crous PW. Cytospora: an important genus of canker pathogens. Stud Mycol 2024;109:323-402. [DOI]
25. Lee JH, Kim YS, Park JT, Ten LN, Lee DH, Jung HY. Increasing incidence of apple Valsa canker and predominance of Cytospora mali in Gyeongsangbuk-do, South Korea. Res Plant Dis 2024;30:325-34. [DOI]
26. Yan C, Hao H, Sha S, Wang Z, Huang L, Kang Z, Wang L, Feng H. Comparative assessment of habitat suitability and niche overlap of three Cytospora species in China. J Fungi 2024;10:38. [DOI]
27. Zhu H, Pan M, Bezerra JDP, Tian C, Fan X. Discovery of Cytospora species associated with canker disease of tree hosts from Mount Dongling of China. MycoKeys 2020;62:97-121.
[DOI]