Tae-Gyeong Kim1, Song-Woon Nam1, Seong-Keun Lim1, Chang-Gi Back2, Seung-Yeol Lee1,3, Leonid N. Ten3, and Hee-Young Jung1,3*
1Department of Plant Medicine, Kyungpook National University, Daegu 41566, Korea
2Department of Environmental Horticulture and Landscape Architecture, Environmental Horticulture, Dankook University, Cheonan 31116, Korea
3Institute of Plant Medicine, Kyungpook National University, Daegu 41566, Korea
*Correspondence to heeyoung@knu.ac.kr
Korean Journal of Mycology (Kor J Mycol) 2025 March, Volume 53, Issue 1, pages 11-20.
https://doi.org/10.4489/kjm.2025.53.1.2
Received on January 15, 2025, Revised on March 09, 2025, Accepted on March 12, 2025, Published on Mar 31, 2025.
Copyright © The Korean Society of Mycology.
This is an Open Access article which is freely available under the Creative Commons Attribution-NonCommercial 4.0 International License (CC BY-NC) (https://creativecommons.org/licenses/by-nc/4.0/).
In 2020, a fungal isolate, designated KNUF-20-112, was obtained from a female rhinoceros beetle (Allomyrina dichotoma) collected in Yangpyeong, Gyeonggi-do, Korea. The isolate formed white, glassy colonies with smooth margins on 4% malt extract/0.5% yeast extract agar. The hyphae measured 6.48–11.24 µm in width and appeared expanded and rigid. Arthroconidia were rectangular in shape, measuring 5.23–9.81 × 13.25–21.41 µm. These morphological characteristics closely matched those of Magnusiomyces magnusii CBS 108.12T. Phylogenetic analyses based on concatenated sequences of the internal transcribed spacer regions (ITS) and large subunit (LSU) gene confirmed the similarity of strain KNUF-20112 with M. magnusii. To our knowledge, this is the first study to document M. magnusii in Korea.
Magnusiomyces, Multilocus sequence analysis, Unreported fungi
The genus Magnusiomyces was established by Zender in 1925, and M. ludwigii was initially designated as its type species [1]. However, subsequent taxonomic revisions based on molecular phylogenetics have redefined the genus, and Magnusiomyces magnusii is now recognized as the type species [2–4]. This genus comprises ascomycetous yeasts and yeast-like fungi characterized by the production of arthroconidia. Species in this group were historically classified under sexual genera, such as Dipodascus, Galactomyces, and Magnusiomyces, and asexual genera, including Geotrichum and Saprochaete [5]. The taxonomy of Magnusiomyces has undergone significant revisions, particularly following the implementation of the principle of “one fungus, one name”, introduced in 2011 by the International Code of Nomenclature for algae, fungi, and plants [6–8]. Under this principle, both the sexual and asexual states of fungi are now unified under a single genus name to reduce taxonomic confusion. Consequently, species previously classified under the asexual genus Saprochaete were reclassified into Magnusiomyces based on molecular phylogenetic studies and nomenclatural priorities [4]. The reclassification of several Saprochaete species into the genus Magnusiomyces has been a significant outcome of recent taxonomic revisions. For example, Saprochaete capitata, an opportunistic pathogen in immunocompromised patients, was reclassified as Magnusiomyces capitatus, whereas Saprochaete clavata, known for its association with nosocomial fungemia outbreaks, is now recognized as Magnusiomyces clavatus [4]. These changes were driven by multilocus sequence analysis (MLSA), which has become a cornerstone of fungal phylogenetics. MLSA has been crucial for resolving the taxonomy of Magnusiomyces. By integrating sequences from loci such as the internal transcribed spacer (ITS) regions and large subunit (LSU) ribosomal RNA, MLSA has played an instrumental role in revising the taxonomy of Magnusiomyces and related genera. This approach has facilitated the identification of novel species and clarified their close evolutionary relationships [2,4,9]. Moreover, it has confirmed that Magnusiomyces species form a distinct monophyletic clade, clearly separated from related genera, such as Geotrichum and Galactomyces [4]. Currently, the genus Magnusiomyces comprises 17 species, including M. capitatus, M. clavatus, M. ingens, M. magnusii, M. ovetensis, M. spicifer and M. tetrasperma [2,3], and several newly recognized species, such as M. chiloensis, M. fungicola, M. gigas [4]. Magnusiomyces species are distinguished based on their asexual and sexual morphological features [3,4]. In its asexual state, colonies are typically white, farinose to hairy, and dry in texture. They consist of true hyphae that branch at acute angles, with acuminate apices, and disarticulate into arthroconidia as the primary reproductive structures. Sympodial or annellidic conidiogenesis may occur in some species; however, chlamydospores are generally absent. In its sexual state, reproduction involves the formation of gametangia on the opposite sides of the hyphal septa. The gametangia fuse to form broadly ellipsoidal hyaline asci, which typically contain four smooth-walled ascospores surrounded by slime sheaths. Ascospores are released after rupture of the ascus wall, facilitating dispersal across various environments.
In this study, we extended our efforts to uncover indigenous Korean fungal species associated with insects, deviating from more commonly explored sources, such as soil and plants. The isolated strain KNUF-20-112 was identified as a member of the genus Magnusiomyces through morphological and molecular analyses. This provides valuable insights into the geographic distribution and ecological diversity of this genus.
In 2020, a female rhinoceros beetle (Allomyrina dichotoma) was collected from Yangpyeong, Gyeonggido, Korea (37°28’03.7″N 127°32’30.3″E) and transported to the laboratory for further analysis. The insect was sterilized by rinsing twice with sterile distilled water and once with 70% ethanol to eliminate surface contaminants. After sterilization, each specimen was ground using a sterile hand grinder to obtain a homogenized sample. The homogenate was suspended in 10 mL of sterile distilled water, vortexed thoroughly, serially diluted, and spread onto yeast extract agar (YEA) plates. YEA plates were incubated at 22°C for 10 days to allow for the development of fungal colonies. After incubation, single fungal colonies were isolated and transferred to fresh YEA plates for further purification under identical conditions. Several fungal strains were successfully isolated and preliminarily identified by sequencing their ITS regions. Among these isolates, strain KNUF-20-112 was identified as a promising candidate as a novel indigenous species in Korea. This strain was then selected for detailed morphological and molecular phylogenetic analyses. A stock culture of strain KNUF-20-112 (NIBRFGC000507834) was deposited as a metabolically inactive culture at the National Institute of Biological Resources (NIBR).
After 10 days of incubation at 22℃ on 4% malt extract/0.5% yeast extract agar (MEYA) [3], the morphological features of isolate KNUF-20-112, including the color, shape, and size, were recorded. Fungal structures were examined under a light microscope (BX-50; Olympus, Tokyo, Japan).
Total genomic DNA was extracted from the fungal mycelia of strain KNUF-20-112 cultured on YEA plates using a HiGene Genomic DNA Prep Kit (BIOFACT, Daejeon, South Korea), according to the manufacturer’s instructions. ITS regions and partial sequences of the 28S rDNA LSU gene were amplified using the ITS1F/ITS4 and LR0R/LR7 primer pairs, respectively [10–12]. The PCR products were purified using ExoSAP-IT PCR Product Cleanup Reagent (Thermo Fisher Scientific, Waltham, MA, USA) and sequenced by Macrogen Sequencing Service (Macrogen, Seoul, Korea). The sequences of the amplified ITS and LSU regions were deposited in GenBank under accession numbers LC859407 and LC859408, respectively.
The ITS and LSU sequences of strain KNUF-20-112 were compared with reference sequences retrieved from the GenBank database of the National Center for Biotechnology Information (NCBI) using the Basic Local Alignment Search Tool. For the phylogenetic analysis, the sequences of the ITS regions and LSU gene were concatenated, and trees were generated using neighbor-joining (NJ), maximum likelihood (ML), and maximum parsimony (MP) methods following the Kimura model [13]. This analysis was performed using MEGA 7 software [14], with bootstrap values calculated from 1,000 replicates to ensure reliability. Reference sequences from the NCBI GenBank database are listed in Table 1.
Table 1. List of species used in phylogenetic analysis along with their GenBank accession numbers
| Species | Strain | GenBank accession numbers | |
|---|---|---|---|
| ITS | LSU | ||
| Geotrichum macrosporus | CBS 259.82T | OP765501 | U40121 |
| Magnusiomyces capitatus | CBS 162.80 | KF984490 | MK834537 |
| Magnusiomyces capitatus | ENCB-HI-834 | MN832904 | MN833644 |
| Magnusiomyces chiloensis | CBS 8187T | OP782220 | MK834538 |
| Magnusiomyces clavatus | CBS 425.71 | KF984489 | KU301161 |
| Magnusiomyces fungicola | CBS 625.85T | OQ586269 | MK834540 |
| Magnusiomyces gigas | CBS 126.76 | AY838940 | MK834547 |
| Magnusiomyces ingens | CBS 521.90T | AY788323 | MK834529 |
| Magnusiomyces japonicus | CBS 100158T | OP778226 | OP821151 |
| Magnusiomyces magnusii | CBS 108.12T | OP821148 | OP821150 |
| Magnusiomyces magnusii | KNUF-20-112 | LC859407 | LC859408 |
| Magnusiomyces ovetensis | CBS 192.55T | OP779246 | MK834533 |
| Magnusiomyces paraingens | CBS 517.90T | OQ586271 | MK834541 |
| Magnusiomyces quercus | CBS 750.85 | OQ586275 | MK834544 |
| Magnusiomyces saccharophilus | CBS 252.91LT | OP779247 | MK834545 |
| Magnusiomyces siamensis | DMKU-GTSP8-14 | MZ322898 | MN460329 |
| Magnusiomyces spicifer | CBS 244.85 | OQ586264 | MK834534 |
| Magnusiomyces starmeri | CBS 780.96T | OP778227 | MK834535 |
| Magnusiomyces suaveolens | CBS 152.25T | AY788291 | MK834546 |
| Magnusiomyces tetraspermus | CBS 765.70T | OQ586266 | MK834536 |
ITS: internal transcribed spacer regions; LSU: 28S rDNA large subunit.
TType strain; LT Lectotype strain.
The strain isolated in this study is indicated in boldface.
After 10 days of incubation at 22℃ on MEYA, the colony diameter of strain KNUF-20-112 ranged from 27.35 to 28.75 mm (Fig. 1A and 1B), closely resembling that of the reference species M. magnusii CBS 108.12 (25 mm). The colony appeared glassy and lobed, with smooth margins, consistent with the reference species. The hyphae of strain KNUF-20-112 were expanding and stiff, with main branches measuring 6.48–11.24 µm in width (7–12 µm in CBS 108.12) and branching at acute angles in a penicillate manner (Fig. 1C). The lateral branches were 4.21–8.38 µm wide (4–7 µm CBS 108.12) and disarticulated into rectangular arthroconidia. The arthroconidia measured 5.23–9.81 × 13.25–21.41 µm (Fig. 1D and 1E), slightly larger than those of the reference species (4–7 × 10–18 µm), but within an acceptable range of variation. Some KNUF-20-112 arthroconidia exhibited annellations at one or both ends, which is a characteristic feature of M. magnusii. These results confirmed that the morphological characteristics of strain KNUF-20-112 closely aligned with those of the reference species M. magnusii CBS 108.12T (Table 2).
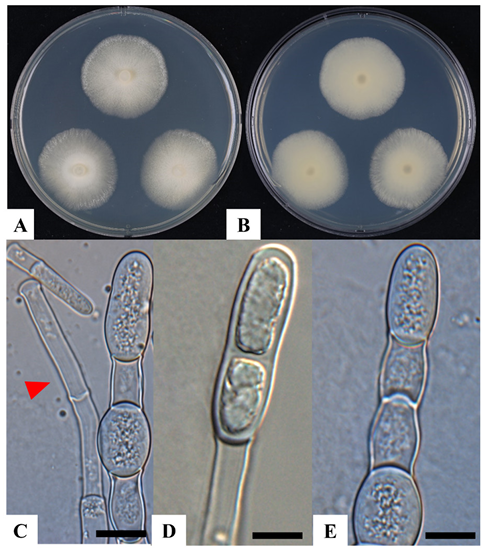
Fig. 1. Cultural and morphological characteristics of Magnusiomyces magnusii KNUF-20-112. A, B: Front and reverse view of colony grown on 4% malt extract/0.5% yeast extract agar (MEYA) after 10 days at 22℃; C: hyphae (indicated by arrowhead); D, E: arthroconidia. Scale bars: C–E = 10 µm.
Table 2. Comparison of morphological characteristics of KNUF-20-112 with reference species Magnusiomyces magnusii CBS 108.12T
| Characteristics | Magnusiomyces magnusii KNUF-20-112a | Magnusiomyces magnusii CBS 108.12b | |
|---|---|---|---|
| Colony | Size (diam) | MEYA: 27.35–28.75 mm in10 days at 22℃ | MEYA: 25 mm in 10 days at 20–22℃ |
| Shape | MEYA: glassy, smooth margin, lobed | MEYA: glassy, smooth margin, lobed | |
| Color | MEYA: white | MEYA: white | |
| Hyphae | Shape | expanding; stiff; branches at acute angles | expanding; stiff; branches at acute angles |
| Wide (µm) | main branches: 6.48–11.24; lateral branches: 4.21–8.38 | main branches: 7–12; lateral branches: 4–7 | |
| Arthroconidia | Shape | rectangular | rectangular |
| Size (µm) | 5.23–9.81 × 13.25–21.41 | 4–7 × 10–18 |
MEYA: 4% malt extract/0.5% yeast extract; diam: diameter.
aFungal strain used in this study; bSource of description [3].
The amplicons generated from the ITS and LSU regions of strain KNUF-20-112 were 591 and 1024 bp in length, respectively. The ITS sequence of this isolate showed 100.00% identity with that of M. magnusii strain CBS 108.12 (OP821148) and 99.31% identity with that of strain CBS 234.85 (OQ586262). Additionally, strain KNUF-20-112 demonstrated a close genetic relationship with several strains of M.gigas, including AUMC 10789 (100.00% similarity, KY495757) and CBS 126.76 (98.96% similarity, OQ586270). A relatively high degree of similarity was also observed between strain KNUF-20-112 and other Magnusiomyces species, including M. suaveolens CBS 152.25 (97.03% similarity, MK834546), M. fragrans LY19 (97.03% similarity, AB499021), M. japonicus CBS 100158 (96.93% similarity, OP778226), and M. tetraspermus CBS 765.70 (96.55% similarity, OQ586266). Based on the similarity of the LSU gene sequences, several strains of M. magnusii were identified as the closest phylogenetic relatives of strain KNUF-20-112, including CBS 108.12 (99.85% similarity, OP821150), NRRL Y-17563 (99.81% similarity, JQ689070), and CBS 234.85 (99.62% similarity, MK834532). Additionally, the isolate demonstrated close genetic relationships with M. suaveolens CBS 152.25 (99.34% similarity, MK834546), M. gigas CBS 126.76 (99.25% similarity, MK834547), M. japonicus CBS 100158 (98.49% similarity, OP821151), M. tetraspermus CBS 765.70 (98.40% similarity, MK834536), M. fungicola CBS 625.85 (97.56% similarity, MK834540), and Magnusiomyces saccharophilus CBS 252.91 (97.37% similarity, MK834545). These results indicate that strain KNUF-20-112 is most closely related to M. magnusii based on the ITS and LSU sequences. However, several other Magnusiomyces species displayed similarity values that exceeded the threshold commonly used for species-level differentiation, highlighting the limitations of relying on a single genetic locus for identification. To address this, MLSA was performed using concatenated ITS and LSU sequences, following the same approach recently applied to revise the genus Magnusiomyces and to describe five new species in the closely related genus Geotrichum [4]. The phylogenetic analysis using the NJ algorithm based on concatenated ITS and LSU sequences showed that strain KNUF-20-112 shared phylogenetic characteristics consistent with those of M. magnusii. Moreover, similar tree topologies were generated using the ML and MP methods, as indicated by the filled circles in Fig. 2, further confirming the phylogeny of the isolate. The combined morphological and phylogenetic evidence supported the identification of KNUF-20-112 as a strain of M. magnusii. To the best of our knowledge, this is the first study documenting this fungal species in Korea.
The genus Magnusiomyces exhibits significant ecological diversity, with species isolated from many natural and anthropogenic sources across different geographic regions [4]. Numerous species are closely associated with arboreal environments. For example, M. magnusii, the type species of the genus, has been isolated from the exudate of oaks (Quercus alba), reflecting its ecological role in tree-associated habitats [3]. Similarly, M. japonicus was recovered from the exudate of a tree, whereas M. quercus was obtained from the slime flux of a red oak in Canada, emphasizing the association of these fungi with tree exudates and decaying woody substrates [2]. Some Magnusiomyces species have adapted to both aquatic and semiaquatic environments. For example, M. saccharophilus has been isolated from a bog pool in Germany, demonstrating its ability to thrive in water-rich habitats. This finding underscores the ability of this genus to persist in diverse ecological niches, possibly because of specific physiological adaptations that enable its survival in such environments. Other species inhabit the substrates associated with decaying organic matter. Magnusiomyces starmeri and M. spicifer have been recovered from rotting cacti in Southern Arizona and the USA, respectively, underscoring their roles as decomposers in arid and semi-arid ecosystems [3]. Additionally, M. suaveolens was isolated from brewery water, illustrating its occurrence in industrial environments containing nutrient-rich substrates. In South Africa, M. ingens and M. paraingens have been isolated from wine cells, suggesting their potential involvement in fermentation [4]. Some species are also associated with anthropogenic environments. For example, M. siamensis has been isolated from food waste in Thailand, indicating its potential role in organic waste degradation [9]. Clinical environments have also yielded notable isolates, including M. clavatus, recovered from human lung tissue in the USA [15], and M. capitatus, isolated from bovine mastitis-associated milk in the UK [16]. These findings highlight the importance of these species as opportunistic pathogens in immunocompromised individuals. Magnusiomyces species are distributed globally. Reports include isolates from North America (e.g., M. spicifer and M. starmeri), Europe (e.g., M. suaveolens and M. saccharophilus), Asia (e.g., M. siamensis), Africa (e.g., M. ingens), and South America (e.g., M. clavatus) [2,3,9]. Its global presence reflects the ecological versatility and adaptability of this genus [4]. The isolation of strain KNUF-20-112, identified as M. magnusii, from a female rhinoceros beetle (Allomyrina dichotoma) in Korea represents a significant and novel finding for this genus. To date, no species within Magnusiomyces have been directly isolated from insects [2,4]. This discovery highlights the potential of insects, such as A. dichotoma, to serve as reservoirs for Magnusiomyces species. As this species has not been previously documented in Korean ecosystems, further studies are necessary to investigate its distribution, ecological roles, and potential interactions with other microorganisms in the local environment.
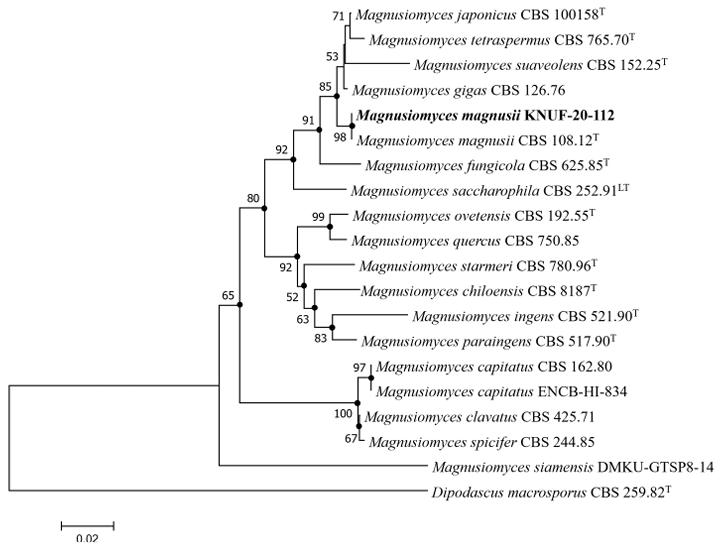
Fig. 2. Neighbor-joining phylogenetic tree based on the concatenated sequences of internal transcribed spacer (ITS) regions and 28S rDNA large subunit (LSU) gene showing the phylogenetic position of strain KNUF-20-112 among Magnusiomyces species. Bootstrap values greater than 50% (based on 1,000 replications) are shown at branch points. The filled circles indicate that the corresponding nodes were also recovered in the trees generated using the maximum likelihood and maximum parsimony algorithms. The isolated strain is indicated in bold. Dipodascus macrosporus CBS 259.82T were used as an out group. Bar, 0.020 substitutions per nucleotide position.
Magnusiomyces comprise species with diverse biological activities that have significant implications for biotechnology, medicine, and ecology. Several species are clinically important owing to their opportunistic pathogenicity. For example, M. capitatus and M. clavatus are associated with systemic infections, such as fungemia and endocarditis in immunocompromised patients, underscoring their medical relevance [17,18]. Many Magnusiomyces species exhibit remarkable enzymatic capabilities, particularly for the breakdown of complex organic compounds. For example, M. spicifer SPB2 exhibits high cell-bound lipase production and tolerance to elevated methanol concentrations, highlighting its potential for biodiesel production via an environmentally friendly conversion process [19]. Other species of this genus also have notable potential for biofuel production. Among these, M. magnusii is an exceptionally versatile organism with robust lipolytic activity and isobutanol-production capabilities, making it an attractive candidate for industrial applications, such as biodiesel production and biofuel synthesis [20].
The isolation of the domestic strain KNUF-20-112, identified as M. magnusii, in Korea is a significant milestone in local research. This strain offers a convenient and sustainable resource for further exploration of the biotechnological potential of this species for various industrial applications. Future studies could leverage KNUF-20-112 to advance renewable energy production, enzyme technology, and other innovative bioprocesses tailored to regional needs.
The authors declare that they have no potential conflicts of interest.
This work was supported by a grant from the National Institute of Biological Resources, funded by the Ministry of Environment of the Republic of Korea (NIBR202002104).
1. Zender J. Sur la classification des Endomycétacées. Bull Soc Bot Genève 1925;17:272-302.
2. de Hoog GS, Smith MT. Ribosomal gene phylogeny and species delimitation in Geotrichum and its teleomorphs. Stud Mycol 2004;50:489-515.
3. de Hoog GS, Smith MT. Magnusiomyces Zender (1977). In: Kurtzman C, Fell JW, Boekhout T, editors. The yeasts, a taxonomic study. 5th ed. Amsterdam: Elsevier; 2011. p. 565-74. [DOI]
4. Zhu HY, Shang YJ, Wei XY, Groenewald M, Robert V, Zhang RP, Li AH, Han PJ, Ji F, Li JN, et al. Taxonomic revision of Geotrichum and Magnusiomyces, with the descriptions of five new Geotrichum species from China. Mycology 2024;15:400-23. [DOI]
5. Kurtzman C, Fell JW, Boekhout T. The yeasts: a taxonomic study. 5th ed. Amsterdam: Elsevier; 2011.
6. Hawksworth D. A new dawn for the naming of fungi: impacts of decisions made in Melbourne in July 2011 on the future publication and regulation of fungal names. MycoKeys 2011;1:7-20. [DOI]
7. Taylor JW. One Fungus = One Name: DNA and fungal nomenclature twenty years after PCR. IMA fungus 2011;2:113-20. [DOI]
8. Turland NJ, Wiersema JH, Barrie FR, Greuter W, Hawksworth DL, Herendeen PS, Knapp S, Kusber WH, Li DZ, Marhold K, et al. (eds.) International Code of Nomenclature for algae, fungi, and plants (Shenzhen Code) adopted by the Nineteenth International Botanical Congress Shenzhen, China, July 2017. Regnum Vegetabile no. 159. Glashütten: Koeltz Botanical Books; 2018. [DOI]
9. Dudhat J, Sakpuntoon V, Angchuan J, Kaewwichian R, Srisuk N. Magnusiomyces siamensis sp. nov., a yeast-like fungus isolated from food waste. Int J Syst Evol Microbiol 2022;72:005435. [DOI]
10. Gardes M, Bruns TD. ITS primers with enhanced specificity for basidiomycetes-application to the identification of mycorrhizae and rusts. Mol Ecol 1993;2:113-8. [DOI]
11. White TJ, Bruns T, Lee S, Taylor J. Amplification, and direct sequencing of fungal ribosomal RNA genes for phylogenetics. In: Innis MA, Gelfand DH, Sninsky JJ, White TJ, editors. PCR protocols: a guide to methods and applications. New York: Academic Press; 1990. p. 315-22. [DOI]
12. Vilgalys R, Hester M. Rapid genetic identification and mapping of enzymatically amplified ribosomal DNA from several Cryptococcus species. J Bacteriol 1990;172:4238-46. [DOI]
13. Kimura M. A simple method for estimating evolutionary rates of base substitutions through comparative studies of nucleotide sequences. J Mol Evol 1980;16:111-20. [DOI]
14. Kumar S, Stecher G, Tamura K. MEGA7: Molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol Biol Evol 2016;33:1870-4. [DOI]
15. Akıncı B, Atay D, Yenigürbüz FD, Akçay A, Akar O, Öztürk G. Fatal disseminated Magnusiomyces clavatus infection with rash in pediatric acute lymphoblastic leukemia: a case report. Diagn Microbiol Infect Dis 2024;108:116148. [DOI]
16. Bosaeed M, Alshehri RA, Albarrak DA, Sharif T, Alghamdi M, Alsunidy AA. An unexpected opportunist: Magnusiomyces capitatus infection in an immunocompetent patient. Med Mycol Case Rep 2024;45:100663. [DOI]
17. Noster J, Koeppel MB, Desnos-Olivier M, Aigner M, Bader O, Dichtl K, Göttig S, Haas A, Kurzai O, Pranada AB, et al. Bloodstream infections caused by Magnusiomyces capitatus and Magnusiomyces clavatus: epidemiological, clinical, and microbiological features of two emerging yeasts species. Antimicrob Agents Chemother 2022;66:e0183421. [DOI]
18. Tanuskova D, Horakova J, Svec P, Bodova I, Lengerova M, Bezdicek M, Poczova M, Koppl J, Kolenova A. First case of invasive Magnusiomyces capitatus infection in Slovakia. Medical Mycol Case Rep 2017;16:12-5. [DOI]
19. Srimhan P, Hongpattarakere T. Scale-up lipase production and development of methanol tolerant whole-cell biocatalyst from Magnusiomyces spicifer SPB2 in stirred-tank bioreactor and its application for biodiesel production. Catalysts 2023;13:617. [DOI]
20. Kurylenko OO, Ruchala J, Dmytruk KV, Abbas CA, Sibirny AA. Multinuclear yeast Magnusiomyces (Dipodascus, Endomyces) magnusii is a promising isobutanol producer. Biotechnol J 2020;15:1900490. [DOI]