1Department of Plant Medicine, Jeonbuk National University, Jeonju 54896, Korea
2Research Center for Plant Medicine, Jeonbuk National University, Jeonju 54896, Korea
3Department of Forestry, Environment, and Systems, Kookmin University, Seoul 02707, Korea
4Division of Environmental Science and Ecological Engineering, Korea University, Seoul 02841, Korea
*Correspondence to iychoi@jbnu.ac.kr, hdshin@korea.ac.kr
Korean Journal of Mycology (Kor J Mycol) 2025 September, Volume 53, Issue 3, pages 145-150.
https://doi.org/10.4489/kjm.2025.53.3.1
Received on June 02, 2025, Revised on July 21, 2025, Accepted on August 12, 2025, Published on September 30, 2025.
Copyright © The Korean Society of Mycology.
This is an Open Access article which is freely available under the Creative Commons Attribution-Non-Commercial 4.0 International License (CC BY-NC) (https://creativecommons.org/licenses/by-nc/4.0/).
Cercospora mirabilis was first described in 1917 as a leaf-spot fungus on Mirabilis jalapa from the USA. Despite global distribution of plant, this fungus has only been recorded in six countries, including Korea. Although the genus Cercospora was recently revised using advanced molecular phylogenetic approaches, C. mirabilis was not included in that study. Moreover, no validated cultures of this fungus were found in any known authentic culture collection. Therefore, nucleotide sequences for taxonomic purposes and information regarding its phylogeny are lacking. To fill these gaps, six samples demonstrating leaf spot symptoms on M. jalapa were collected from various parts of Korea and examined for morphological and molecular phylogenetic characterization of C. mirabilis. Two newly obtained monoconidial isolates were deposited in an authentic culture collection in Korea. This study provides the molecular phylogeny of C. mirabilis based on multigene sequence analyses, confirming that it is a distinct species within the genus Cercospora.
Cercospora flagellaris, Nyctaginaceae, Pseudocercospora oxybaphi
Mirabilis jalapa L., known as ʻmarvel of Peru’ or ʻfour o’clock flower’, belongs to the family Nyctaginaceae and is native to the arid tropical regions of the North, Central, and South American continents, including Mexico, Guatemala, Chile, and Peru. This plant has been introduced for ornamental purposes and has become naturalized in tropical, subtropical, and temperate regions worldwide [1]. Commercial cultivars are available with various flower colors. Despite its global distribution and worldwide planting as an ornamental plant, records of phytopathogenic fungi on this plant are scarce. A powdery mildew fungus, Pseudoidium nyctaginacearum (Hosag.) U. Braun & R.T.A. Cook from eight countries [2] and the leaf spot fungus Cercospora mirabilis Tharp from six countries are the two major pathogens of M. jalapa [3–5]. Phytopathogens that are rare and confined to a specific region include Pseudocercospora oxybaphi (Ellist & Halst.) U. Braun & Crous from the USA and Phytophthora mirabilis Galindo & H.R. Hohl from Mexico, with some minor records requiring confirmation [6].
Cercospora mirabilis was first described in 1917 on M. jalapa in Texas, USA [7]. Additional records of this fungus have been obtained from China, India, France, Sierra Leone, and Korea [6]. However, C. mirabilis could not be included in a recent extensive study on Cercospora species that used advanced molecular phylogenetic approaches, presumably because of the lack of living cultures [8]. Although most literature sources have listed C. mirabilis without a morphological description, Kim and Shin [9] have provided observations and illustrations of this fungus. However, the morphology and ecology of this fungus have not been sufficiently studied. To the best of our knowledge, no living cultures of C. mirabilis are available in culture collections [https://wfcc.info/]. Therefore, this study aimed to provide information on the morphological characteristics and molecular sequence of C. mirabilis on M. jalapa and to ensure the deposition of the first fungal isolate in an authentic culture collection center.
During the course of this study, six specimens were collected from various parts of Korea and preserved at the Korea University Herbarium (Seoul, Korea) for morphological examination. These were as follows: KUS-F14452 (17 Oct 1997, Seodun-dong, Suwon), F15599 (28 Oct 1998, Wabu village, Namyangju), F317159 (22 Oct 1999, Deokso farm, Namyangju), and F32007 (4 Oct 2020, Naksan Park, Seoul). To obtain monoconidial isolates of C. mirabilis, two additional samples were collected from Seoul and Jeonju in October 2024 and later deposited as voucher specimens in the fungorium of Jeonbuk National University (accession numbers JBNU-F0570 and JBNU-F0575).
Detailed morphological characteristics of the fungus were observed using fresh samples under an Olympus BX50 microscope (Olympus, Tokyo, Japan). Photomicrographs were captured using a Zeiss AX10 microscope equipped with an AxioCam MRc5 camera (Carl Zeiss, Oberkochen, Germany). For fungal isolation, conidia were collected from young lesions, mounted in a drop of sterile water, and streaked onto 2% water agar (WA; Junsei, Tokyo, Japan) plates supplemented with 100 mg/L streptomycin sulfate. The inoculated plates were then incubated at 25℃. After two days, the germinating conidia were transferred onto 2% potato dextrose agar (PDA; Difco, Detroit, MI, USA) plates to obtain a pure culture.
Leaf spots were amphigenous, scattered, distinct, small, circular to subcircular, mostly not exceeding 4 mm in diameter, rarely reaching up to 10 mm when confluent, initially pale brown to tan, and later the center became dull gray due to plentiful sporulation, with or without blackish-brown margins (Fig. 1A). Caespituli were amphigenous. Stromata were small to medium, composed of swollen hyphal cells, brown to dark brown, subglobular to angular, and 20–42 μm in diameter. (Fig. 1B). Conidiophores were 4 to 18 in a divergent fascicles, arising from substomatal stromata or emerging through the cuticle, olivaceous brown to grayish brown throughout, usually uniform in width, straight to mildly curved, rarely 2 to 3 times geniculate, 1–3(–4)-septate, and 50–140 × 4–5 μm; conidial scars were large, 2–3 μm wide, conspicuous, apical or on shoulders of conidiogenous cells (Fig. 1C). Conidia were solitary, filiform to cylindrical, straight to slightly curved, 2–10-septate, hyaline, non-constricted at the septa, obtuse at the apex, subtruncate to obtuse at the base, 24–110 × 3.5–5.5 μm; hila were conspicuously thickened, darkened, and non-protuberant (Fig. 1D). These characteristics are consistent with previous descriptions of C. mirabilis [9–11].
The culture characteristics of C. mirabilis have not been described previously. Two isolates obtained in this study showed the same growth pattern on PDA. Two-week-old colonies grown on PDA at 25℃ were 25–30 mm in diameter, with an entire margin. The obverse surface was irregularly folded with prominent network of ridges, pale olivaceous to pinkish color with clear undulate, dark pink margin, the reverse of colony was dark olive, with blood red margin (Fig. 1E). The two isolates were deposited in the Korean Agricultural Culture Collection (KACC) under the accession numbers KACC 410991 and KACC 410992.
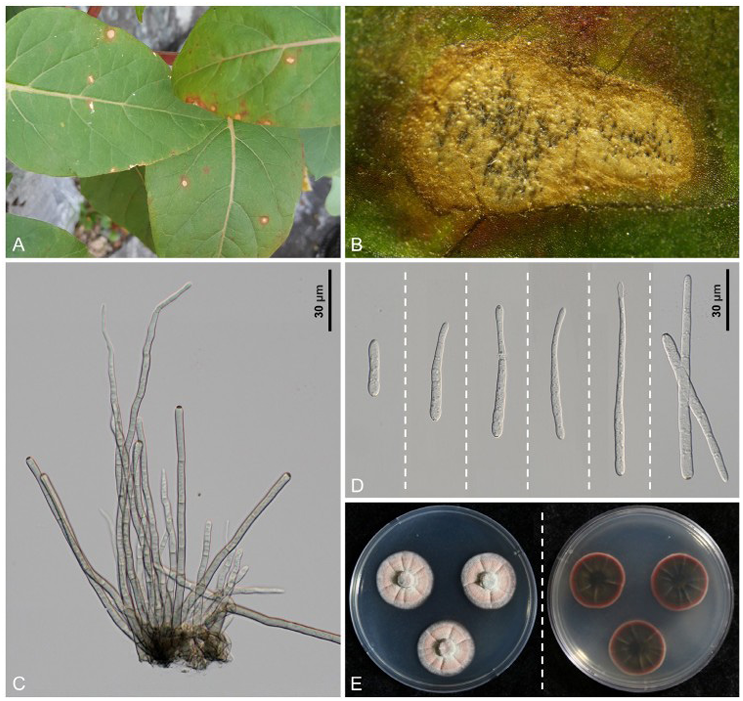
Fig. 1. Cercospora mirabilis found on Mirabilis jalapa. A: Leaf spots caused by C. mirabilis. B: Closeup of a leaf lesion with abundant sporulation. C: Conidiophores with stroma. D: Conidia. E: The colony appearance (obverse and reverse) of C. mirabilis after 14 days of growth on potato dextrose agar (PDA) at 25℃.
For molecular phylogenetic analyses, the nucleotide sequences of the internal transcribed spacer (ITS) region of the rDNA and four protein-coding genes, partial actin (actA), partial calmodulin (cmdA), partial histone H3 (his3), and partial translation elongation factor 1-alpha (tef1), were amplified and sequenced using primer pairs outlined by Groenewald et al. [8]. As primers CAL-228F and CAL-737R failed to amplify the cmdA gene; therefore, a new primer set was developed during our study: C-CAL-F (5ʹ-TTCTCCCTCTTCGTACGTAC-3ʹ) for forward, and C-CAL-R (5ʹ-CATGGTAAGGAATTCGGGG3ʹ) for the reverse. Conventional PCR for the amplification of the cmdA gene was conducted in the following procedures: initial denaturation at 94℃ for 2 min; 94℃ for 30 sec of denaturation, 52℃ for 30 sec of annealing, followed by extension at 72℃ for 30 sec for 40 cycles; and final extension at 72℃ for 5 min. The PCR products were sequenced using the same primers by Bionics (Seoul, Korea). The obtained raw sequences were inspected, assembled, and deposited in GenBank under the accession numbers PV389800 and PV389803 for ITS, PV409659 and PV409660 for actA, PV409661 and PV409662 for cmdA, PV409663 and PV409664 for his3, and PV409665 and PV409666 for tef1. The dataset for each gene was generated using sequences of closely related Cercospora spp. retrieved from the GenBank database and aligned individually in Molecular Evolutionary Genetics Analysis version 11 (MEGA 11) using the Multiple Sequence Comparison by Log-Expectation (MUSCLE) tool [12]. Subsequently, the prepared datasets were concatenated into a single multigene dataset of ITS+tef1+actA+cmdA+his3 using Sequence Matrix [13]. Septoria provencialis (CPC 12226) was designated as an outgroup [8]. The phylogenetic tree was constructed based on the maximum parsimony method in PAUP*4.0a using a heuristic search option with a ‘tree-bisection’ algorithm [14]. The robustness of the internal branches was assessed using bootstrap analysis (BS) with 1,000 replicates and scores higher than 70% were assigned to relevant branches. Tree scores including tree length (TL = 931), consistency index (CI = 0.4480), retention index (RI = 0.5537), and rescaled consistency index (RC = 0.3265) were also calculated. The final alignment consisted of 40 sequences and 1,701 characters, of which 204 (11.99%) were variable and parsimony-uninformative, whereas 213 (12.52%) were informative for parsimony analysis.
The results of the BLASTn search in GenBank were 100% identical to Cercospora capsici (GU214653, KT193659) for the ITS region. The coding sequence regions showed the highest percentage (100%) identity with the sequences determined as C. cf. flagellaris for actA (JX143114, DQ835121, etc.), his3 (JX142621, JX142624, etc.), cmdA (JX142866, DQ835145, etc.), and tef1 sequences (JX143360, JX143362, etc.). In the phylogenetic tree (Fig. 2), the two isolates obtained in this study formed a clade with the C. cf. flagellaris group. Phytolacca americana L. (syn: P. decandra L.) was the original host of C. flagellaris Ellis and G. Martin [15]. In our analyses, an isolate of C. flagellaris found on P. americana (CPC10124; South Korea) was also placed in the same group with C. (cf.) flagellaris sequences and our isolates. Because of the lower BS value (58%) of the cf. flagellaris branch, which indicates weak support for grouping, it was not shown in the tree.
Despite the phylogenetic proximity of C. mirabilis and C. flagellaris, clear and consistent morphological differences were observed. In Chupp’s description of C. flagellaris [11], conidial septation was reported as indistinct, conidia were up to 120(–280) µm long, not exceeding 4 µm in width, and conidiophores were up to 300 µm long. In addition, the morphological characterization of C. flagellaris on P. americana given by Kim & Shin indicates conidial septation range as 4–16, conidial length up to 130(–200) µm, conidial width reaching 4.5 µm, and conidiophores up to 280 µm long [9,10]. Therefore, C. flagellaris could be differentiated from our isolates by its longer and narrower conidia, with a greater septation range and longer conidiophores. In conclusion, although C. mirabilis is phylogenetically close to the C. cf. flagellaris group, it can be recognized as a taxonomically distinct species based on consistent and stable morphological differences.
Historically, two species of Cercospora have been described that were found on Mirabilis species.
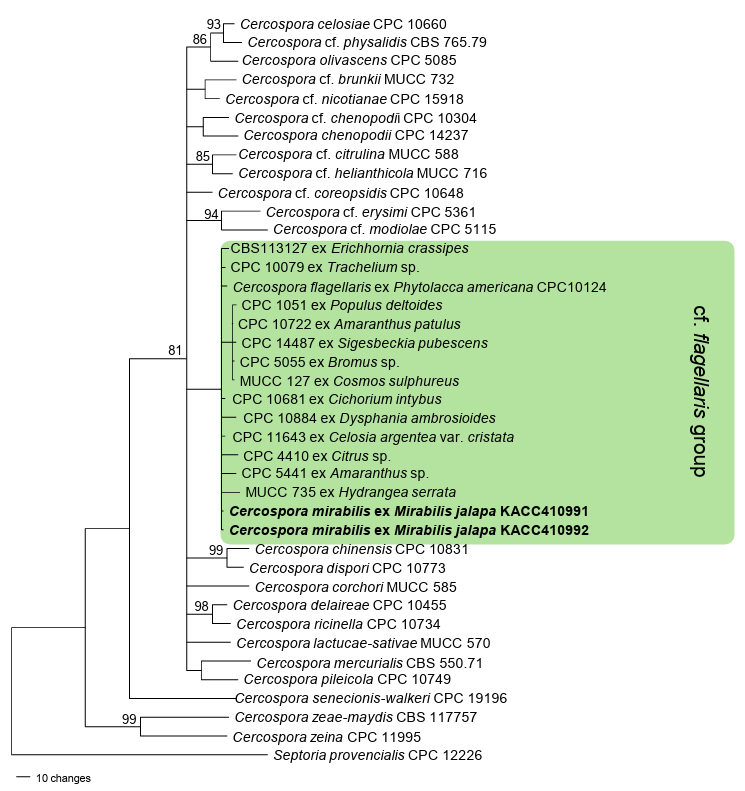
Fig. 2. A maximum parsimony tree for C. mirabilis was generated based on a combined dataset of internal transcribed spacer (ITS), translation elongation factor 1-alpha (tef1), actin (actA), calmodulin (cmdA), and histone H3 (his3) sequences. Forty sequences were included in the analysis. Septoria provencialis (CPC12226) was selected as an outgroup. Isolates obtained in this study are shown in bold. Bootstrap values (≥70%) are indicated for related branches.
Among these, C. oxybaphi Ellis & Everh. was first described in 1888 as a leaf spot fungus on M. nyctaginea (syn. Oxybaphus nyctagineus) in the USA. This fungus was transferred to Pseudocercospora; therefore, P. oxybaphi (Ellis & Everh.) U. Braun & Crous is currently accepted [3]. Thus, C. mirabilis is the only member of the genus Cercospora known to be associated with Mirabilis and closely related plant taxa. As stated above, the morphological characteristics of C. mirabilis have been described and illustrated in Korea [9]. Nevertheless, photomicrographs of stromata, conidiophores, and conidia showing taxonomic value are presented for the first time in this study. The phylogenetic placement of C. mirabilis, inferred from multigene sequence analyses, is revealed here for the first time. This study thoroughly characterized C. mirabilis using both morphological and molecular data, contributing to its diagnostic purposes and enhancing our understanding of its phylogenetic relationship within the genus Cercospora.
The authors declare no conflicts of interest.
This research was supported by the Global-Learning & Academic Research Institution for Master’s·PhD students and Postdocs (LAMP) Program of the National Research Foundation of Korea (NRF) grant funded by the Ministry of Education (No. RS-2024-00443714).
1. Liya FI, Yasmin MF, Chowdhury NS, Charu TK, Fatema IB. Mirabilis jalapa: A review of ethno and pharmacological activities. Adv Med Plant Res 2021;9:1-10.
2. Braun U, Cook RTA. Taxonomic manual of the Erysiphales (Powdery Mildews). CBS Biodivers Series. Utrecht: CBS-KNAW Fungal Biodiversity Centre; 2012.
3. Crous PW, Braun U. Mycosphaerella and its anamorphs: 1. Names published in Cercospora and Passalora. Utrecht: Centraalbureau voor Schimmelcultures; 2003.
4. Shin HD, Braun U. Notes on Korean Cercosporae and allied genera (III). Mycotaxon 2000;74:105-18. [DOI]
5. Guo YL, Xu L. Studies on Cercospora and allied genera in China XI. Mycosystema 2002;21:181-4.
6. Farr DF, Rossman AY. Fungal Databases [Internet]. U.S. National Fungus Collections. Washington, D.C.: Agricultural Research Service, U.S. Department of Agriculture [cited 2025 April 2]. Available from: https://nt.ars-grin.gov/fungaldatabases/
7. Tharp BC. Texas parasitic fungi. Mycologia 1917;9:105-24. [DOI]
8. Groenewald JZ, Nakashima C, Nishikawa J, Shin HD, Park JH, Jama AN, Groenewald M, Braun U, Crous PW. Species concepts in Cercospora: Spotting the weeds among the roses. Stud Mycol 2013;75:115-70. [DOI]
9. Kim JD, Shin HD. Taxonomic studies on Cercospora and allied genera in Korea (IX). Kor J Mycol 1999;27:211-9.
10. Shin HD, Kim JD. Cercospora and allied genera from Korea. Suwon: National Institute of Agricultural Science and Technology; 2001.
11. Chupp C. A monograph of the fungus genus Cercospora. Ithaca: Published by the author; 1954.
12. Tamura K, Stecher G, Kumar S. MEGA11: Molecular evolutionary genetics analysis version 11. Mol Biol Evol 2021;38:3022-7. [DOI]
13. Vaidya G, Lohman DJ, Meier R. SequenceMatrix: Concatenation software for the fast assembly of multi-gene datasets with character set and codon information. Cladistics 2011;27:171-80. [DOI]
14. Swofford DL. PAUP: Phylogenetic analyses using parsimony (and other methods) 4.0b10. Sunderland: Sinauer Associates. 2002.
15. Ellis JB, Martin GB. New species of North American fungi. Am Nat 1882;16:1001-4.