1Department of Plant Medicine, Kyungpook National University, Daegu 41566, Korea
2Community Engagement for Food and Agriculture Development in Rwanda, Kigali, Rwanda
3Institute of Plant Medicine, Kyungpook National University, Daegu 41566, Korea
*Correspondence to leesy1123@knu.ac.kr
Korean Journal of Mycology (Kor J Mycol) 2025 September, Volume 53, Issue 3, pages 151-162.
https://doi.org/10.4489/kjm.2025.53.3.2
Received on July 01, 2025, Revised on July 23, 2025, Accepted on August 12, 2025, Published on September 30, 2025.
Copyright © The Korean Society of Mycology.
This is an Open Access article which is freely available under the Creative Commons Attribution-Non-Commercial 4.0 International License (CC BY-NC) (https://creativecommons.org/licenses/by-nc/4.0/).
This study reports the discovery of two previously unreported fungal strains belonging to Hypocreales, designated KNUF-20-085 and KNUF-20-NI014, from soil samples in Korea. Phylogenetic analyses based on the concatenated nucleotide sequences of the internal transcribed spacer regions and partial sequences of the translation elongation factor 1-alpha and second-largest subunit of RNA polymerase II genes placed the strains within the genus of Trichoderma. We investigated the strains cultural features on potato dextrose agar, malt extract agar, corn meal agar, and synthetic nutrient-poor agar, and their morphological characteristics through microscopic observation. For strain KNUF-20-085, cultural and morphological characteristics showed a high similarity with those of Trichoderma panacis SYPF 8050T. For strain KNUF-20-NI014, cultural and morphological characteristics were similar to those previously reported for T. brevicompactum CBS 109720T. The phylogenetic analysis supported these affiliations, with strains KNUF-20-085 and KNUF-20-NI014 clustering with T. panacis and T. brevicompactum, respectively. This study represents the first documentation of T. panacis and T. brevicompactum in Korea.
Morphology, Phylogeny, Trichoderma brevicompactum, Trichoderma panacis
Hypocreales is an order within the phylum Ascomycota and the class Sordariomycetes. Members of this order are typically recognized by their bright, lightly pigmented fruiting bodies [1]. Hypocreales comprises 15 families, including Hypocreaceae, Bionectriaceae, Nectriaceae, and Clavicipitaceae [2]. Within Hypocreaceae, 17 genera have been identified, among which Trichoderma is prominent [2,3]. Trichoderma was first proposed as a genus by C. H. Persoon in 1794 based on material collected in Germany [4]. Due to changes in the International Code of Nomenclature, the genus Trichoderma has been proposed for conservation over its teleomorph, Hypocrea. Thus, all species bearing both the names Hypocrea and Trichoderma, as well as those described solely under Hypocrea, have been officially transferred to the genus Trichoderma [5], which now includes more than 400 species, including the type species T. fuliginoides [2]. The genus Trichoderma is commonly found in and isolated from a diverse range of habitats, such as soils, moist wood, tropical forest trees, mushrooms, bracket fungi in forests, and even water-damaged buildings [5,6]. Species in this genus are typically characterized by rapid growth, the production of bright green masses of conidia, a repetitively branched conidiophore structure, and their role as plant symbionts [7,8]. Another well-known characteristic of many Trichoderma species is their antifungal or plant-growthstimulating activities, along with the production of unique enzymes and secondary metabolites [3]. These features have made them exploitable as biological control agents against fungal phytopathogens, with some isolates currently used in commercially available applications [9–11]. Due to their enzymatic versatility and secondary metabolite production, species of Trichoderma also play critical roles in ecological nutrient cycling, decomposition, and plant–microbe interactions [3], and they are increasingly valued in sustainable agriculture, particularly as eco-friendly alternatives to chemical fungicides [3]. Traditionally, taxonomic studies of Trichoderma species were based on morphological and physiological characteristics [12]. However, as the number of described species has grown, distinguishing them solely through morphological observation has become difficult due to high degrees of similarity among many species. As a result, DNA sequence analysis has become the new standard in fungal phylogenetics and systematics. Phylogenetic analyses combining the sequences of the internal transcribed spacer (ITS) regions and the translation elongation factor 1-alpha (TEF1) and second-largest subunit of RNA polymerase II (RPB2) genes are now widely used to study phylogenetic relationships within Trichoderma and to reveal taxonomic diversity [13]. Although the genus Trichoderma has been widely studied worldwide, only 33 species have been reported from environmental samples in Korea [14–19]. Therefore, the two Trichoderma species isolated for the first time from Korean soil in this study contribute to expanding our current understanding of fungal diversity in the region. In this paper, we document and describe the morphological and phylogenetic characteristics of these strains.
Fungal strains were isolated using the plate dilution method from soil samples collected in Pohang-si (36°11’05.8″N 129°23’03.4″E) and Dokdo (37°14’28.9″N, 131°51’54.5″E), Gyeongbuk province in Korea. Strains KNUF-20-085 and KNUF-20-NI014 were selected from numerous fungal strains for further morphological and molecular phylogenetic analysis. Stock cultures of strains KNUF-20-085 and KNUF20-NI014 were deposited in the National Institute of Biological Resources (NIBR) as metabolically inactive cultures under accession numbers NIBRFGC000507845 and NIBRFGC000507835, respectively.
Cultural and micromorphological characteristics were studied using different cultural media for each strain. Cultural characteristics, including colony texture, color, size, and shape, were examined using potato dextrose agar (PDA; Difco, Detroit, MI, USA) and corn meal agar (CMA; Difco, Detroit, MI, USA) for both strains, while malt extract agar (MEA; Difco) was used exclusively for strain KNUF-20-085 and synthetic nutrient-poor agar (SNA; MBcell, Seoul, Korea) was used exclusively for strain KNUF-20NI014, with an incubation temperature of 20℃ for all cultures [20,21]. Micromorphological features were observed under a light microscope (BX-50; Olympus, Tokyo, Japan).
Total genomic DNA was extracted from fungal mycelia cultured on PDA using a HiGeneTM Genomic DNA Prep Kit (BIOFACT, Daejeon, Korea) according to the manufacturer’s instructions. Molecular identification was conducted by analyzing the ITS regions with partial RPB2 and partial TEF1 gene sequences, which were amplified using the primer pairs ITS1F/ITS4, fRPB2-5f/fRPB2-7cr, and EF1728F/TEF1LLErev, respectively [22–25]. The amplified PCR products were purified using the EXOSAPIT PCR Product Cleanup Reagent (Thermo Fisher Scientific, Waltham, MA, USA) and sequenced by SolGent (Daejeon, South Korea). The obtained sequences of strains KNUF-20-085 and KNUF-20-NI014 were deposited in the National Center for Biotechnology Information (NCBI) GenBank database (Table 1).
Table 1. GenBank accession numbers of sequences used for the phylogenetic analyses in this study
테이블
ITS: internal transcribed spacer regions; TEF1: translation elongation factor 1-alpha; RPB2: RNA polymerase II subunit.
TType strain.
The newly generated sequences are indicated in bold.
The ITS regions, TEF1, and RPB2 gene sequences of strains KNUF-20-085 and KNUF-20-NI014 were aligned with reference sequences retrieved from the NCBI GenBank database. Ambiguous regions were deleted from the alignments, and evolutionary distance matrices for the neighbor-joining (NJ) algorithm were calculated using the Kimura two-parameter model [26,27]. Phylogenetic relationships were inferred via the topology of trees generated using the NJ method in MEGA11 software with 1,000 bootstrap replications [28].
Trichoderma panacis S.Y. Liu, T. Yuan Zhang, Ying Yu & Yi X. Zhang, Int J Syst Evol Microbiol 70 (5): 3165 (2020) [MB#824197]
Specimen collection: Cheongha-myeon, Pohang-si, Gyeongbuk province, Korea (36°11’05.8″N, 129°23’03.4″E), isolated from soil.
Description: Colonies on PDA reached 32–36 mm in diameter after 72 h of culturing at 20℃. Mycelia were hyaline and whitish, with aerial hyphae, wavy margins, and downy to finely floccose surfaces (Fig. 1A). A coconutlike odor was detected, with no diffusing pigment observed. On MEA, colonies reached 15–22 mm in diameter after 72 h of culturing at 20℃. Colonies were dense and whitish, with well-defined margins, aerial hyphae, and downy to floccose surfaces (Fig. 1B). A coconut-like odor was detected, with no diffusing pigment observed. On CMA, colonies reached 14–18 mm in diameter after 72 h of culturing at 20℃. Colonies were hyaline and thin, with aerial hyphae, and slightly wavy margins (Fig. 1C). Hyphae were aerial, radial, and sometimes inconspicuous. Conidiophores were Verticillium-like, substituted by phialides singly or in whorls (Fig. 1D). Phialides were produced in a terminal cluster (Fig. 1E and F). Conidia were ellipsoidal, smooth, yellowish green to green, and 4–5 × 2.3–3.2 µm (n = 40) (Fig. 1G). Chlamydospores were not observed.
Notes: Comparing the strains T. panacis KNUF-20-085 and SYPF 8050ᵀ, both strains exhibit similar cultural characteristics on PDA, MEA, and CMA media, and share morphological traits such as smooth, ellipsoidal conidia formed at the ends of Verticillium-like conidiophores (Table 2) [20].
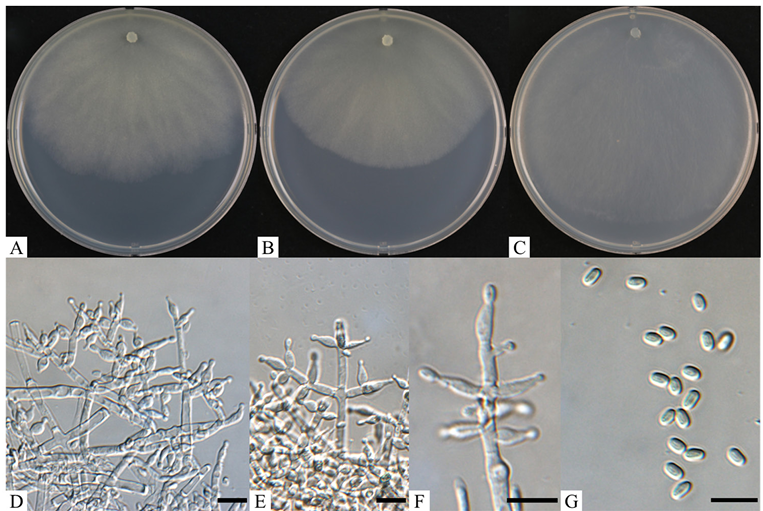
Fig. 1. Cultural and morphological characteristics of Trichoderma panacis KNUF-20-085. A–C: Front views of colonies after 7 d at 20℃ on potato dextrose agar, malt extract agar, and corn meal agar, respectively; D: conidiophores and phialides with hyphae; E: phialides at the ends of branched hyphae; F: phialides; G: conidia. Scale bars = 10 µm.
Table 2. Cultural and morphological comparison of Trichoderma panacis KNUF-20-085 with closely related Trichoderma strains
테이블
PDA: potato dextrose agar; MEA: malt extract agar; CMA: corn meal agar.
TType strain; aFungal strain studied in this research; bSource of descriptions [20]; cSource of description [36]; dSource of description [37].
Amplification of the ITS regions, RPB2, and TEF1 gene of strain KNUF-20-085 yielded 573, 922, and 892 bp fragments, respectively. The ITS regions of strain KNUF-20-085 showed a 99.3% similarity with that of Trichoderma panacis SYPF 8050T, a 99.1% similarity with those of T. erinaceum strains CEN1420 and CEN1421, and a 99.0% similarity with that of T. koningii APSAC 01. For the partial TEF1 gene sequence, strain KNUF-20-085 showed a 98.8% similarity with T. panacis SYPF 8050T, 98.6% similarities with T. erinaceum strains DUCC15708 and CEN1558, and a 96.0% similarity with T. atroviride HNG4-3. In the case of the partial RPB2 gene sequence, strain KNUF-20-085 showed 100% similarity with T. erinaceum DUCC15708, a 99.8% similarity with T. panacis SYPF 8050T, and a 97.0% similarity with T. songyi SFC20130926-S001 (Fig. 2). In the NJ phylogenetic tree generated using the concatenated ITS regions, RPB2, and TEF1 gene sequences, strain KNUF-20-085 clustered together with T. panacis SYPF 8050T. Thus, based on the morphological and phylogenetic analyses, strain KNUF20-085 was identified as T. panacis.
Trichoderma brevicompactum G.F. Kraus, C.P. Kubicek & W. Gams, Mycol. 96 (5): 1063 (2004) [MB#487780]
Specimen collection: Dokdo, Ulleung-gun, Gyeongbuk province, Korea (37°14’28.9″N, 131°51’54.5″E), isolated from soil.
Description: Colonies on PDA reached 44–50 mm in diameter after 72 h of culturing at 20℃. Mycelia were f ilamentous, white, and broad, with concentric rings, entire margins, and yellowish green to green conidia (Fig. 3A). On CMA, colonies reached 42–47 mm in diameter after 72 h of culturing at 20℃. Mycelia were white, with scant arial hyphae, forming yellowish green conidia (Fig. 3B). On SNA, colonies reached 33–36 mm in diameter after 72 h of culturing at 20℃. Mycelia were white, with scant arial hyphae and yellowish green conidia (Fig. 3C). Hyphae were white, terminal, verticillate, and fertile, with the distal parts of some hyphae forming conidiogenous hyphal cells commonly with one to three conidiogenous loci (Fig. 3D). Conidiophore branches arose from the surface and were verticillate, fertile, and terminal, with phialides (Fig. 3E). Phialides were produced in a dense terminal cluster, ampulliform, and slightly enlarged in the middle (Fig. 3F). Conidia were subglobose, yellowish green to green, smooth, and 3.0–4.2 × 2.5–3.3 µm (n = 40) (Fig. 3G). Chlamydospores developed in older cultures and were subglobose and intercalary or terminal (Fig. 3H).
Notes: Comparing the T. brevicompactum KNUF-20-NI014 and MA 3296T, both strains exhibit similar cultural characteristics on PDA, SNA, and CMA media, and share morphological traits such as subglobose, smooth, green conidia formed at the ends of verticillate conidiophores (Table 3) [21].
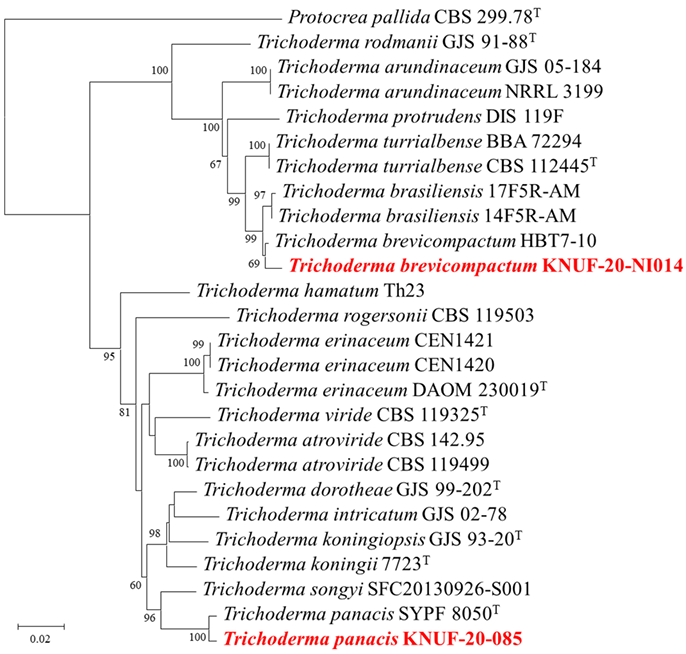
Fig. 2. Neighbor-joining phylogenetic tree based on the concatenated sequences of internal transcribed spacer (ITS) regions, RNA polymerase II subunit B (RPB2) and translation elongation factor 1-alpha (TEF1) gene showing the phylogenetic position of strains KNUF-20-085 and KNUF-20-NI014 among Trichoderma species. Bootstrap values greater than 60% (percentage of 1,000 replications) are shown at branching points. The strain isolated in this study is in bold and red. The tree was rooted using Protocrea pallida CBS 299.78T as an out-group. Bar, 0.02 substitutions per nucleotide position.
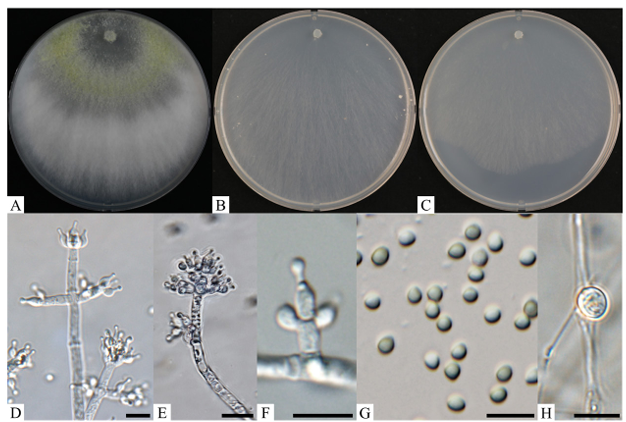
Fig. 3. Cultural and morphological characteristics of Trichoderma brevicompactum KNUF-20-NI014. A–C: Front views of colonies after 7 d at 20℃ on potato dextrose agar, corn meal agar, and synthetic nutrient-poor agar, respectively; D: conidiophores and phialides with conidia; E: conidiophores and phialides; F: phialides; G: conidia; H: chlamydospore. Scale bars = 10 µm.
Table 3. Cultural and morphological comparison of Trichoderma brevicompactum KNUF-20-NI014 with closely related Trichoderma strains
테이블
PDA: potato dextrose agar; CMA: corn meal agar; SNA: synthetic nutrient-poor agar.
TType strain; aFungal strain studied in this research; bSource of descriptions [21]; cSource of description [35].
Amplification of ITS regions, RPB2, and TEF1 gene of strain KNUF-20-NI014 yielded 559, 980, and 527 bp fragments, respectively. The ITS regions of strain KNUF-20-NI014 showed 100% similarities with those of various strains of T. brevicompactum, including strain CEN510, CEN1071, 100% similarity with that of T. turrialbense CBS 112445, and 100% similarity with that of T. brasiliensis 14F5R-AM. The partial TEF1 gene sequence of strain KNUF-20-NI014 showed 98.1–98.6% similarities with those of various T. brevicompactum strains, including strains DAOM 233362, 27RCS, GJS 04-381, and CBS 112444, 95.0% similarities with those of T. brasiliensis strains 14F5R-AM and 17F5R-AM, and a 92.7% similarity with that of T. turrialbense BBA 72294. For the partial RPB2 gene sequence, strain KNUF-20-NI014 showed 100% similarities with various T. brevicompactum strains, including strains BF06, HZA7, and HBG1-1, a 99.7% similarity with T. brasiliensis 17F5R-AM, and a 98.2% similarity with T. turrialbense BBA 72294. A phylogenetic tree was constructed using the NJ method based on the concatenated ITS regions, RPB2, and TEF1 gene sequences (Fig. 2). In this phylogenetic tree, strain KNUF-20-NI014 clustered closely with T. brevicompactum HBT7-10. For the phylogenetic analysis, the type strain MA 3296T was not used due to the lack of an RPB2 gene sequence, and thus, another T. brevicompactum strain was used. Overall, the cultural, morphological, and phylogenetic analyses collectively identified strain KNUF-20-NI014 as T. brevicompactum.
Since the genus was first established in 1794 [4], Trichoderma species have been isolated from a wide range of sources [5,6]. In this study, strains KNUF-20-085 and KNUF-20-NI014, both isolated from soil samples in Korea, were identified as T. panacis and T. brevicompactum, respectively. As there are no previous records of T. panacis and T. brevicompactum from Korea, this study represents the first official report of these species in the country. Various Trichoderma species are well-recognized for their roles as biocontrol agents due to their production of bioactive secondary metabolites and enzymes, as well as their ability to suppress mycotoxin production [29]. To date, a number of studies have addressed the usage of the secondary metabolites of the T. brevicompactum complex as antibiotics [30]. The term “complex” here refers to a group of closely related species within Trichoderma that are morphologically similar to T. brevicompactum but genetically distinct from each other and often difficult to distinguish using traditional identification methods. Trichoderma brevicompactum is reported to produce trichothecene (trichodermin), which exhibits inhibitory activity against fungal pathogens like Rhizoctonia solani and Botrytis cinerea [31]. In addition, a study of the antagonistic effects of T. brevicompactum against fungal plant pathogens revealed significant inhibition of Fusarium oxysporum growth and development with disruption of physiological structures and spore formation [32]. Therefore, the antibiotics and metabolites of strain KNUF-20-NI014 warrant further research, as they may have potential future applications as commercially viable bioprotective agents against fungal diseases. In the case of T. panacis, there are no reports of antibiotic activities at the time of this study. However, some closely related species, such as T. erinaceum and T. atroviride, are known to produce secondary metabolites exhibiting antifungal activity [33–35]. Therefore, further investigation of T. panacis is warranted to determine whether it possesses similar biosynthetic pathways for the production of secondary metabolites and antibiotics that may contribute to its use as a biological control agent.
The discovery of the newly reported species isolated in this study, T. brevicompactum and T. panacis, contributes to our understanding of domestic biodiversity and its conservation. Their descriptions may benefit future research in ecology and the development of human-related and industrial applications of fungus-derived products. As Trichoderma is noteworthy for its species’ important roles in biocontrol and abilities to persist in soils through crop rotations and in intercropping systems, conducting systematic studies in this genus will be essential for informing future scientific applications.
No potential conflict of interest was reported by the authors.
This work was supported by a grant from the National Institute of Biological Resources (NIBR), funded by the Ministry of Environment (MOE) of the Republic of Korea [NIBR202002104].
1. Chaverri P, Samuels GJ. Hypocrea/Trichoderma (Ascomycota, Hypocreales, Hypocreaceae): Species with green ascospores. Stud Mycol 2003;48:1-116. [DOI]
2. Wijayawardene NN, Hyde KD, Dai DQ, Sánchez-García M, Goto BT, Saxena RK, Erdoğdu M, Selçuk F, Rajeshkumar KC, Aptroot A, et al. Outline of fungi and fungus-like taxa-2021. Mycosphere 2022;13:53-453. https://doi.org/10.5943/mycosphere/13/1/2 [DOI]
3. Samuels GJ. Trichoderma: Systematics, the sexual state, and ecology. Phytopathol 2006;96:195-206. https://doi.org/10.1094/phyto-96-0195 [DOI]
4. Persoon CH. Disposita methodical fungorum. Romers Neues Mag Bot 1794;1:81-128.
5. Rossman AY, Seifert KA, Samuels GJ, Minnis AM, Schroers HJ, Lombard L, Crous PW, Põldmaa K, Cannon PF, Summerbell RC, et al. Genera in Bionectriaceae, Hypocreaceae, and Nectriaceae (Hypocreales) proposed for acceptance or rejection. IMA Fungus 2013;4:41-51. https://doi.org/10.5598/imafungus.2013.04.01.05 [DOI]
6. Evans HC, Holmes KA, Thomas SE. Endophytes and mycoparasites associated with an indigenous forest tree, Theobroma gileri, in Ecuador and a preliminary assessment of their potential as biocontrol agents of cocoa diseases. Mycol Prog 2003;2:149-60. https://doi. org/10.1007/s11557-006-0053-4 [DOI]
7. Gams W, Bissett J. Morphology and identification of Trichoderma. In: Kubicek CP, Harmann GE, editors. Trichoderma and Gliocladium. London: Taylor and Francis; 2002. p. 3-34.
8. Harman GE, Howell CR, Viterbo A, Chet I, Lorito M. Trichoderma species-opportunistic, avirulent plant symbionts. Nat Rev Microbiol 2004;2:43-56. https://doi.org/10.1038/ nrmicro797 [DOI]
9. Bae H, Roberts DP, Lim HS, Strem MD, Park SC, Ryu CM, Melnick RL, Bailey BA. Endophytic Trichoderma isolates from tropical environments delay disease onset and induce resistance against Phytophthora capsici in hot pepper using multiple mechanisms. Mol Plant Microbe Interact 2011;24:336-51. [DOI]
10. Zeilinger S, Gruber S, Bansal R, Mukherjee PK. Secondary metabolism in Trichoderma chemistry meets genomics. Fungal Biol Rev 2016;30:74-90. https://doi.org/10.1016/ j.fbr.2016.05.001 [DOI]
11. Mironenka J, Różalska S, Soboń A, Bernat P. Lipids, proteins and extracellular metabolites of Trichoderma harzianum modifications caused by 2, 4-dichlorophenoxyacetic acid as a plant growth stimulator. Ecotoxicol Environ Saf 2020;194:110383. https://doi.org/10.1016/ j.ecoenv.2020.110383 [DOI]
12. Shentu X, Zhan X, Ma Z, Yu X, Zhang C. Antifungal activity of metabolites of the endophytic fungus Trichoderma brevicompactum from garlic. Braz J Microbiol 2014;45:248-54. https:// doi.org/10.1590/S1517-83822014005000036 [DOI]
13. Cai F, Druzhinina IS. In honor of John Bissett: Authoritative guidelines on molecular identification of Trichoderma. Fungal Divers 2021;107:1-69. https://doi.org/10.1007/s13225020-00464-4 [DOI]
14. Goh J, Nam B, Lee JS., Mun HY, Oh Y, Lee HB, Chung N, Choi YJ. First report of six Trichoderma species isolated from freshwater environment in Korea. Kor J Mycol 2018;46:213-25. [DOI]
15. Huh N, Jang Y, Lee J, Kim GH, Kim JJ. Phylogenetic analysis of major molds inhabiting woods and their discoloration characteristics. Part 1. Genus Trichoderma. Holzforschung 2011;65:257-63. https://doi.org/10.1515/hf.2011.018 [DOI]
16. Jang S, Jang Y, Kim CW, Lee H, Hong JH, Heo YM, Lee YM, Lee DW, Lee HB, Kim JJ. Five new records of soil-derived Trichoderma in Korea: T. albolutescens, T. asperelloides, T. orientale, T. spirale, and T. tomentosum. Mycobiology 2017;45:1-8. https://doi.org/10.5941/ MYCO.2017.45.1.1 [DOI]
17. Ahn GR, Kim JE, Oh YS, Lee KM, Jin H, Kim MU, Kim JY, Kim SH. Undescribed fungal species of Eupenicillium, Mortierella, and Trichoderma isolated in the vicinity of demilitarized zone in Yeoncheon-gun, Gyeonggi-do. Korea. Kor J Mycol 2018;46:359-67. [DOI]
18. Jang S, Kwon SL, Lee H, Jang Y, Park MS, Lim YW, Kim C, Kim JJ. New report of three unrecorded species in Trichoderma harzianum species complex in Korea. Mycobiology 2018;46:177-84. https://doi.org/10.1080/12298093.2018.1497792 [DOI]
19. Goh J, Oh Y, Park YH, Mun HY, Park S, Cheon W. Isolation and characterization of previously undescribed seventeen fungal species belonging to the order Hypocreales in Korea. Kor J Mycol 2022;50:1-29. [DOI]
20. Liu SY, Yu Y, Zhang TY, Zhang MY, Zhang YX. Trichoderma panacis sp. nov., an endophyte isolated from Panax notoginseng. Int J Syst Evol Microbiol 2020;70:3162-6. https://doi. org/10.1099/ijsem.0.004144 [DOI]
21. Degenkolb T, Dieckmann R, Nielsen KF, Gräfenhan T, Theis C, Zafari D, Chaverri P, Ismaiel A, Brückner H, von Döhren H, et al. The Trichoderma brevicompactum clade: A separate lineage with new species, new peptaibiotics, and mycotoxins. Mycol Prog 2008;7:177-219. https://doi.org/10.1007/s11557-008-0563-3 [DOI]
22. White TJ, Bruns T, Lee S, Taylor J. Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics. In: Innis MA, Gelfand DH, Sninsky JJ, et al., editors. PCR protocols: A guide to methods and applications. New York: Academic Press; 1990. p. 315-22. [DOI]
23. Gardes M, Bruns TD. ITS primers with enhanced spec ificity for basidiomycetes-application to the identifica tion of mycorrhizae and rusts. Mol Ecol 1993;2:113-8. https://doi.org/10.1111/ j.1365-294x.1993.tb00005.x [DOI]
24. O’Donnell K, Cigelnik E. Two divergent intragenom ic rDNA ITS2 types within a monophyletic lineage of the fungus Fusarium are nonorthologous. Mol Phylogenet Evol 1997;7:103-16. https://doi.org/10.1006/mpev.1996.0376 [DOI]
25. Jaklitsch WM, Komon M, Kubicek CP, Druzhinina IS. Hypocrea voglmayrii sp. nov. from the Austrian Alps represents a new phylogenetic clade in Hypocrea/Trichoderma. Mycologia 2005;97:1365-78. https://doi.org/10.3852/mycologia.97.6.1365 [DOI]
26. Saitou N, Nei M. The neighbor-joining method: A new method for reconstructing phylogenetic trees. Mol Biol Evol 1987;4:406-25. https://doi.org/10.1093/oxfordjournals.molbev.a040454 [DOI]
27. Kimura M. A simple method for estimating evolutionary rates of base substitutions through comparative studies of nucleotide sequences. J Mol Evol 1980;16:111-20. https://doi. org/10.1007/BF01731581 [DOI]
28. Kumar S, Stecher G, Li M, Knyaz C, Tamura K. MEGA X: Molecular evolutionary genetics analysis across computing platforms. Mol Biol Evol 2018;35:1547-9. https://doi.org/10.1093/ molbev/msy096 [DOI]
29. Braun, H, Woitsch L, Hetzer B, Geisen R, Zange B, Schmidt-Heydt M. Trichoderma harzianum: Inhibition of mycotoxin producing fungi and toxin biosynthesis. Int J Food Microbiol 2018;28:10-6. https://doi.org/10.1016/j.ijfoodmicro.2018.04.021 [DOI]
30. Degenkolb T, Gräfenhan T, Nirenberg HI, Gams W, Brückner H. Trichoderma brevicompactum complex: Rich source of novel and recurrent plant-protective polypeptide antibiotics (peptaibiotics). J Agric Food Chem 2006;54:7047-61. https://doi.org/10.1021/ jf060788q [DOI]
31. Shentu X, Zhan X, Ma Z, Yu X, Zhang C. Antifungal activity of metabolites of the endophytic fungus Trichoderma brevicompactum from garlic. Braz J Microbiol 2014;45:248-54. https:// doi.org/10.1590/S1517-83822014005000036 [DOI]
32. Tian L, Zhu X, Guo Y, Zhou Q, Wang L, Li W. Antagonism of rhizosphere Trichoderma brevicompactum DTN19 against the pathogenic fungi causing corm rot in saffron (Crocus sativus L.) in vitro. Front Microbiol 2014;15: 1454670. https://doi.org/10.3389/ fmicb.2024.1454670 [DOI]
33. Siebatcheu EC, Wetadieu D, Youassi OY, Bedine Boat MA, Bedane KG, Tchameni NS, Sameza ML. Secondary metabolites from an endophytic fungus Trichoderma erinaceum with antimicrobial activity towards Pythium ultimum. Nat Prod Res 2023;37:657-62. https://doi.or g/10.1080/14786419.2022.2075360 [DOI]
34. Stracquadanio C, Quiles JM, Meca G, Cacciola SO. Antifungal activity of bioactive metabolites produced by Trichoderma asperellum and Trichoderma atroviride in liquid medium. J Fungi 2020;6:263. https://doi.org/10.3390/jof6040263 [DOI]
35. Kraus GF, Druzhinina I, Gams W, Bissett J, Zafari D, Szakacs G, Koptchinski A, Prillinger H, Zare R, Kubicek CP. Trichoderma brevicompactum sp. nov. Mycologia 2004;96:1059-73. [DOI]
36. Park MS, Oh SY, Cho HJ, Fong JJ, Cheon WJ, Lim YW. Trichoderma songyi sp. nov., a new species associated with the pine mushroom (Tricholoma matsutake). Antonie van Leeuwenhoek 2014;106:593-603. https://doi.org/10.1007/s10482-014-0230-4 [DOI]
37. Bissett J, Szakacs G, Nolan CA, Druzhinina IS, Kullnig-Gradinger CM, Kubicek CP. Seven new taxa of Trichoderma from Asia. Canad J Bot 2003;81:570-86. [DOI]