1Department of Plant Medicine, Jeonbuk National University, Jeonju 54896, Korea
2Research Center for Plant Medicine, Jeonbuk National University, Jeonju 54896, Korea
3Division of Environmental Science and Ecological Engineering, Korea University, Seoul 02841, Korea
*Correspondence to iychoi@jbnu.ac.kr, hdshin@korea.ac.kr
Korean Journal of Mycology (Kor J Mycol) 2025 September, Volume 53, Issue 3, pages 163-170.
https://doi.org/10.4489/kjm.2025.53.3.3
Received on March 18, 2025, Revised on August 12, 2025, Accepted on September 08, 2025, Published on September 30, 2025.
Copyright © The Korean Society of Mycology.
This is an Open Access article which is freely available under the Creative Commons Attribution-Non-Commercial 4.0 International License (CC BY-NC) (https://creativecommons.org/licenses/by-nc/4.0/).
Pseudocercospora broussonetiae is a foliar pathogen that causes leaf spot disease on Broussonetia species in China and Japan. The most recent mention of this fungus dates back to the early 2000s, and unsurprisingly, no molecular phylogenetic data are available to support its differentiation into distinct species. This study is the first to confirm the occurrence of Ps. broussonetiae on B. papyrifera in Korea, characterizing its morphological features and phylogenetic position among other Pseudocercospora species, and providing the first sequence data. Our isolates were largely morphologically consistent with previously reported characteristics of Ps. broussonetiae, although minor variations were observed, particularly in the size of the conidiophores and conidia. A phylogenetic tree constructed from a multigene dataset of the ITS (internal transcribed spacer), actA (actin), tef1 (translation elongation factor 1-alpha), and rpb2 (RNA polymerase II second-largest subunit) confirmed that the Korean isolates were placed in a distinct clade with Ps. davidiicola and apart from other Pseudocercospora species on mulberry hosts. This study provides new insights into the morphology and phylogeny of Ps. broussonetiae within the genus Pseudocercospora.
Cercosporoid fungi, Moraceae, Mycosphaerellaceae, Paper mulberry, Pseudocercospora davidiicola
Pseudocercospora Speg. (Mycosphaerellaceae) is a diverse genus of cercosporoid fungi that is widely distributed in various ecosystems. Members of this genus infect many plants, including angiosperms, gymnosperms, and ferns, as foliar pathogens [1,2]. This genus exhibits high morphological and genetic diversity, which makes species delimitation challenging, particularly because of its close relationship with the Cercospora and Passalora genera [2]. Numerous species remain poorly characterized, with limited morphological descriptions and genetic data, which hinders a comprehensive understanding of their taxonomy and evolution. As new species are continuously discovered, accurate identification is essential for clarifying their taxonomic positions.
A notable example is Pseudocercospora broussonetiae (Chupp & Linder) Y.L. Guo & X.J. Liu (MycoBank no. MB 126469), which is associated with leaf spot disease on Broussonetia L’Hér. ex Vent. (Moraceae). This fungus was first described on B. papyrifera (L.) Vent. in China as Cercospora broussonetiae Chupp & Linder in 1937 [3]. It was subsequently reclassified under the genus Pseudocercospora as Ps. broussonetiae based on the absence of conidiogenous loci (scars) and hila [4]. The last mention of this fungus dates back to the early 2000s; after that, no new reports were made [5]. Currently, B. kaempferi Siebold, B. papyrifera, and Broussonetia sp. in China and B. kazinoki Siebold in Japan are listed as hosts for this pathogen [6]. To date, no molecular phylogenetic data have been available to support its classification. This study aimed to fill these gaps by providing a detailed characterization of fungal morphology and determining the phylogenetic position of this species within the Pseudocercospora genus using isolates collected from Korea.
Two samples of B. papyrifera representing Pseudocercospora leaf spots preserved at the herbarium of Korea University (KUS, Seoul, Korea) and Jeonbuk National University (JBNU, Jeonju, Korea) as KUS-F34412 (3 Nov 2024, Seoul) and JBNU-F0606 (7 Nov 2024, Jeonju) were used in this study.
Morphological examinations were performed using fresh materials under an Olympus BX50 microscope (Olympus, Tokyo, Japan), and photomicrographs were captured using a Zeiss AX10 microscope equipped with an AxioCam MRc5 camera (Carl Zeiss, Oberkochen, Germany). The size of the diagnostic features represented the average of at least 30 measurements taken for each feature. To obtain monoconidial isolates, conidia from young lesions were suspended in sterile water in microcentrifuge tubes. The resulting conidial suspension was then streaked onto 2% water agar (WA; Junsei, Tokyo) plates containing 100 mg/ L of streptomycin sulfate and incubated at 25°C. After 5 days of incubation, the growing colonies were transferred to 2% potato dextrose agar (PDA; Difco, France) plates. The isolated colonies were deposited in the Korean Agricultural Culture Collection (KACC, Rural Development Administration, Wanju, Korea) under accession numbers KACC 411024 (KUS-F34412) and KACC 411021 (JBNU-F0606).
Leaf spots were amphigenous, scattered, angular to irregular, 3–5(–10) mm wide, creamy or necrotic in the center, surrounded by a dark brown margin on the upper surface, and dark brown to light brown on the lower surface due to the fructification (Fig. 1A–B). Caespituli were amphigenous but primarily hypophyllous and solitary (Fig. 1C). The external mycelia were grayish, branched, and septate. Stromata were well-developed, globose, pale brown to dark brown, erumpent, and 25–50 µm in diameter. Conidiophores were (5–)7–17 × 3–4 µm, brown to pale brown, smooth-walled, subcylindrical, straight to slightly flexuous, 0–2-septate, arising from external hyphae in dense numerous fascicles on stromata. Conidiogenous cells were unbranched, pale to light brown, apex obtuse, and inconspicuous scars (Fig. 1DE). Conidia were solitary, cylindrical to obclavate, straight to slightly curved, more or less constricted at the septa, pale olivaceous, guttulate, 2–6-septate, apex obtuse and obconically truncate base, 23–64 × 3.5–4.5 µm, with unthickened and not darkened hila (Fig. 1E–F). Three-week-old colonies grown on PDA at 25°C were 21–25 mm in diameter, the surface was irregularly radiated with a lobate margin, and the upper surface was khaki in the center but olivaceous near the margin, developed moderate aerial hyphae (Fig. 1G). The morphological characteristics of the isolates were consistent with those of Ps. broussonetiae [7].

Fig. 1. Leaf spot disease caused by Pseudocercospora broussonetiae on Broussonetia papyrifera. A: Symptoms of infected plants in the field; B: Leaf spots on the lower surface; C: Fructification of the fungus on the lesion, showing a grayish to brown mass of conidia; D: Conidiophores on a stroma; E: Conidiogenous cells and mass of conidia; F: Conidia; G: Three-week-old colonies of Ps. broussonetiae on potato dextrose agar at 25°C. Scale bar indicates 20 µm.
To confirm the morphology-based identification, the nucleotide sequences of the internal transcribed spacer (ITS1 and ITS2) regions and protein-coding genes, including partial actin (actA), partial translation elongation factor 1-alpha (tef1), and partial DNA-directed RNA polymerase II second-largest subunit (rpb2) of rDNA, were determined from 2-week-old colonies grown on PDA [8,9]. Newly obtained sequences were assembled, tested against reference sequences in GenBank, and submitted to the National Center for Biotechnology Information (NCBI) database (PV249131 and PV249316 for ITS, PV288330 and PV259889 for actA, PV259887 and PV259890 for tef1, and PV259888 and PV259891 for rpb2). A data matrix was created for each gene individually and aligned in MEGA11 using the MUSCLE command or phylogenetic analysis [10]. All four datasets were then concatenated into a single multigene dataset in the order ITS + actA + tef1 + rpb2 using SequenceMatrix [11]. Trochophora simplex (CBS 214744) was selected as an outgroup [8]. Phylogenetic trees were generated in PAUP*4.0a for maximum parsimony (MP) analysis through a heuristic search option using a ‘tree-bisection’ algorithm [12], and in raxmlGUI 2.0.13 for maximum likelihood (ML) analysis based on the GAMMA model with General TimeReversible (GTR) substitution [13]. Bootstrap analyses (BS) were performed with 1000 replications to test the robustness of the branches of the consensus trees; subsequently, ≥70% of the BS values were shown on the related branches. Tree scores, including tree length, consistency index, retention index, and rescaled consistency index were calculated. The final alignment consisted of 35 sequences and 1,848 characters, of which 181 (9.79%) were variable and parsimony-uninformative, whereas 500 (27.01%) were informative for parsimony analysis. The best nucleotide substitution model was determined to be GTR + I + G, using MrModelTest v.2.2. before the Bayesian inference analysis [14]. Markov Chain Monte Carlo analysis of the four chains was performed in parallel using a random tree topology. The heating parameter was set to 0.1, and trees were saved every 20,000 generations until the average standard deviation of the split frequencies reached a stop value of 0.01 in MrBayes v. 3.2.7 [15]. A consensus tree was visualized using FigTree v. 1.4.4 (Institute of Evolutionary Biology, University of Edinburgh, http://tree.bio.ed.ac.uk/software/figtree). Moreover, the calculated Bayesian posterior probabilities are indicated on the nodes of the parsimonious tree (Fig. 2).
The two ITS sequences obtained in this study, each 675/676 bp long, differed by a single nucleotide (cytosine vs. adenine). In addition, they were 100% identical to each other in actA, rpb2, and tef1. The BLASTn search revealed ~99.6% and 99.7% similarity with Pseudocercospora sp. on Macroptilium atropurpureum (OQ793671) for ITS, whereas 100% identity was found with Ps. cornicola (MT711895), Ps. cercidicola (GU384388), and Ps. abeliae (LC599448) for tef1, and Ps. stranvaesiae (LC599623), Ps. prunicola (PP404585), Ps. forsythiae (LC599606), and Ps. naitoi (KX462644) for rpb2. The actA sequences showed 100% identity with those of Pseudocercospora sp. on Populus sp. (KM982614), Ps. cornicola (MT711893), and Ps. paraguayensis (MZ614953). In the MP tree, our two isolates clustered in a clade distinct from Ps. davidiicola (MUCC 296) and Ps. humuli (MUCC 742), and within a clade, they are grouped in a branch with Ps. davidiicola. In the phylogenetic tree, it was difficult to determine whether the Ps. broussonetiae clade represents a monophyletic group. Only one Ps. humuli strain (MUCC 742) and one Ps. davidiicola strain (MUCC 296) have all four gene sequences in the GenBank database. Therefore, it was not possible to add additional Ps. davidiicola and Ps. humuli in this study.
Although our isolates were largely consistent with the previously reported characteristics of Ps. broussonetiae [7], minor variations have been observed, particularly in the size of the conidiophores and conidia. Significant differences were the presence of longer conidiophores (up to 65 µm) and conidia (20.072.5 µm) in the Chinese description. Several factors, such as intraspecific variation (natural variability within species across different regions), environmental influences, and practical differences in methodology, can explain this variation. Moreover, our phylogenetic results indicate a close evolutionary relationship between Ps. davidiicola and Ps. broussonetiae. To determine the distinct traits, we compared the morphologies of the specimens with Ps. davidiicola. According to the description of the type material for the latter, its conidia were longer (15–86 µm) and had more septation in conidia and conidiophores (up to 10-septate) [16] (Table 1).
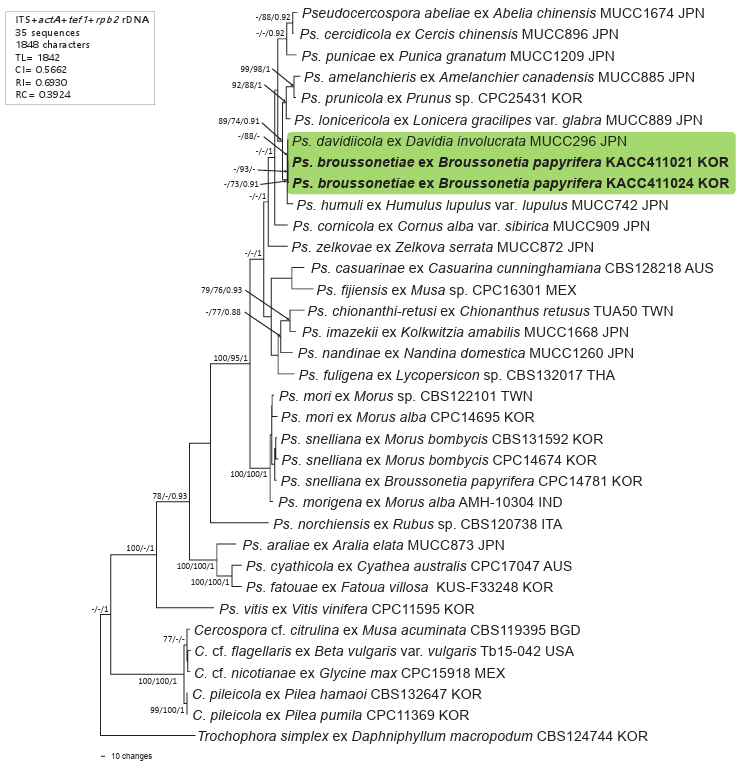
Fig. 2. Maximum parsimony tree of Pseudocercospora broussonetiae on Broussonetia papyrifera generated from a combined multigene dataset of 35 sequences of the ITS + actA + tef1 + rpb2 genes. The isolates obtained in this study are shown in bold. Bootstrap values (>70%) from maximum parsimony and maximum likelihood analyses, as well as posterior probabilities (>90%) from Bayesian inference analysis are given on the relevant branches and nodes, respectively. Tree scores, such as tree length (TL), consistency index (CI), retention index (RI), and rescaled consistency index (RC), are presented in the box on the left side of the Figure. ITS: internal transcribed spacer; actA: actin; tef1: translation elongation factor 1-alpha; rpb2: RNA polymerase II second-largest subunit.
An isolate of Ps. snelliana (Reichert) U. Braun, H.D. Shin, C. Nakash. & Crous on B. papyrifera from Korea (CPC 14781) was included in the phylogenetic analyses of the latest study by Groenewald et al. [17], without any description of this record. This fungus was introduced as a new combination in 2013 based on the type material on Morus alba L. [2]. According to its description, the conidia of Ps. snelliana are dark colored, ellipsoid-ovoid, subcylindrical, distinctly obclavate in shape, and longer (up to 86 μm), conidiophores are longer (up to 100 μm) and up to 12-septate, which are key diagnostic traits to distinguish it from Ps. broussonetiae (Table 1). To assess the evolutionary relationships among isolates, Ps. snelliana and other Psedocercospora species on Morus spp., viz. Ps. mori (Hara) Deighton, and Ps. morigena A. Singh, P.N. Singh & N.K. Dubey were included in the phylogenetic analyses. Our isolates identified as Ps. broussonetiae were distinctly separated from Ps. sneliana and other Pseudocercospora species on Morus spp., supporting its classification.
Table 1. Morphological comparison of Korean Pseudocercospora broussonetiae with other allied Pseudocercospora species
테이블
The genus Broussonetia consists of four species, namely, B. harmandii Gagnep., B. kaempferi, B. monoica Hance, and B. papyrifera, along with one natural hybrid, Broussonetia × kazinoki [18]. Among these, B. papyrifera, commonly known as paper mulberry, is native to Eastern Asian countries, including China, Korea, and Japan, and is cultivated for traditional uses such as papermaking [19]. A finding of Ps. broussonetiae on this plant in Korea represents the spread of the pathogen across East Asia, where traditional papermaking is an important industry.
To the best of our knowledge, this is the first study to confirm the occurrence of Ps. broussonetiae on B. papyrifera in Korea. Detailed morphological features of the fungus, its phylogenetic position among other Pseudocercospora species, and the first sequence data are provided. Consequently, our findings shed light on the morphology and phylogeny of Ps. broussonetiae, revealing its host range, distribution, and taxonomic position within the genus Pseudocercospora.
No potential conflict of interest was reported by the author(s).
This research was supported by the Global-Learning and Academic Research Institute for Master’s PhD students and Postdocs (LAMP) Program of the National Research Foundation of Korea (NRF) grant funded by the Ministry of Education (No. RS-2024-00443714).
1. Guatimosim E, Schwartsburd PB, Barreto RW, Crous PW. Novel fungi from an ancient niche: Cercosporoid and related sexual morphs on ferns. Persoonia 2016;37:106-41. https://doi. org/10.3767/003158516×690934 [DOI]
2. Crous PW, Braun U, Hunter GC, Wingfield MJ, Verkley GJM, Shin HD, Nakashima C, Groenewald JZ. Phylogenetic lineages in Pseudocercospora. Stud Mycol 2013;75:37-114. https://doi.org/10.3114/sim0005 [DOI]
3. Chupp C, Linder DH. Notes on Chinese Cercospora. Mycologia 1937;29:26-33. [DOI]
4. Guo YL, Liu XJ. Studies on the genus Pseudocercospora in China. I. Mycosystema 1989;2:225-40.
5. Braun U, Crous PW, Pons N. Annotated list of Cercospora species (epithets a-b) described by C. Chupp. Feddes Repert 2002;113:112-27. https://doi.org/10.1002/1522239X(200205)113:1/2%3C112::AID-FEDR112%3E3.0.CO;2-H [DOI]
6. Farr DF, Rossman AY. Fungal databases, US national fungus collections, Agricultural Research Service (ARS), United States Department of Agriculture (USDA) [Internet]. Beltsville, MD: USDA; 2025 [cited 2025 Mar 13]. Available from: https://fungi.ars.usda.gov/.
7. Guo YL, Hsieh WH. The genus Pseudocercospora in China. Beijing, China: International Academic Publishers; 1995.
8. Nakashima C, Motohashi K, Chen CY, Groenewald JZ, Crous PW. Species diversity of Pseudocercospora from Far East Asia. Mycol Prog 2016;15:1093-117. https://doi.org/10.1007/ s11557-016-1231-7 [DOI]
9. Choi IY, Abasova L, Park JH, Shin HD. Revisiting Pseudocercospora puerariicola associated with leaf spot on Pueraria montana var. lobata with morphological and molecular-phylogenetic data in Korea. J Plant Pathol 2025;107:675-80. https://doi.org/10.1007/s42161-024-01749-2 [DOI]
10. Tamura K, Stecher G, Kumar S. MEGA11: Molecular evolutionary genetics analysis version 11. Mol Biol Evol 2021;38:3022-7. [DOI]
11. Vaidya G, Lohman DJ, Meier R. SequenceMatrix: Concatenation software for the fast assembly of multi-gene datasets with character set and codon information. Cladistics 2011;27:171-80. https://doi.org/10.1111/j.1096-0031.2010.00329.x [DOI]
12. Swofford DL. PAUP: Phylogenetic analyses using parsimony (*and Other Methods) 4.0a10. Sunderland, MA: Sinauer Associates. 2002.
13. Silvestro D, Michalak I. RaxmlGUI: A graphical front-end for RAxML. Org Divers Evol. 2012;12:335-7. https://doi.org/10.1007/s13127-011-0056-0 [DOI]
14. Nylander JAA. MrModeltest 2.0. Program distributed by the author. Uppsala, Sweden: Uppsala University; 2004.
15. Ronquist F, Teslenko M, van der Mark P, Ayres DL, Darling A, Höhna S, Larget B, Liu L, Suchard MA, Huelsenbeck JP. MrBayes 3.2: Efficient Bayesian phylogenetic inference and model choice across a large model space. Syst Biol 2012;61:539-42. https://doi.org/10.1093/ sysbio/sys029 [DOI]
16. Motohashi K, Araki I, Nakashima C. Four new species of Phyllosticta, one new species of Pseudocercospora, and one new combination in Passalora from Japan. Mycoscience 2008;49:138-46. https://doi.org/10.1007/S10267-007-0395-Z [DOI]
17. Groenewald JZ, Chen YY , Zhang Y, Roux J, Shin HD, Shivas RG, Summerell BA, Braun U, Alfenas AC, Ujat AH, et al. Species diversity in Pseudocercospora. Fungal Syst Evol 2024;13:29-89. https://doi.org/10.3114/fuse.2024.13.03 [DOI]
18. Plants of the World Online. Facilitated by the Royal Botanic Gardens, Kew [Internet]. Richmond, UK: Royal Botanic Gardens, Kew; 2025 [cited 2025 Mar 13]. Available from https://powo.science.kew.org/.
19. Whistler WA, Elevitch CR. Broussonetia papyrifera (paper mulberry). In: Elevitch CR, editor. Traditional trees of Pacific Islands: Their culture, environment, and use. Holualoa, HI: Permanent Agriculture Resources; 2006.