1Department of Plant Medicine, Jeonbuk National University, Jeonju 54896, Korea
2Research Center for Plant Medicine, Jeonbuk National University, Jeonju 54896, Korea
3Plant Quarantine Technology Center, Department of Plant Quarantine, Animal and Plant Quarantine Agency, Gimcheon 39660, Korea
4Division of Environmental Science and Ecological Engineering, Korea University, Seoul 02841, Korea
*Correspondence to iychoi@jbnu.ac.kr, hdshin@korea.ac.kr
Korean Journal of Mycology (Kor J Mycol) 2025 September, Volume 53, Issue 3, pages 193-201.
https://doi.org/10.4489/kjm.2025.53.3.6
Received on August 13, 2025, Revised on September 09, 2025, Accepted on September 09, 2025, Published on September 30, 2025.
Copyright © The Korean Society of Mycology.
This is an Open Access article which is freely available under the Creative Commons Attribution-Non-Commercial 4.0 International License (CC BY-NC) (https://creativecommons.org/licenses/by-nc/4.0/).
Cercospora cf. flagellaris is one of the unresolved species complexes within the genus Cercospora. Despite recent advances in molecular approaches for species delimitation using multigene sequence analysis, the identity of C. flagellaris remains unclear. As new records are accumulated, the complexity of this species group increases, encompassing hosts from a wide range of plant families. Phytolacca americana, a species native to North America and later introduced to East Asia, was the original host of this fungus. Since 1991, leaf spot diseases caused by C. flagellaris have been consistently reported in Korea. In this study, we summarized all isolates identified as C. flagellaris from P. americana in Korea to date, to infer their morphological and molecular phylogenetic identities and to understand the true identity of this fungus. All isolates were morphologically and molecularly examined, illustrated, and their phylogenetic placement was demonstrated in a tree inferred from multigene sequence analyses.
Leaf spot disease, Multigene analysis, Mycosphaerellaceae, Species complex
The genus Cercospora Fresen. ex Fuckel (Mycosphaerellaceae) contains numerous economically and ecologically important plant pathogenic fungi that cause leaf spot disease on various plants, including angiosperms, gymnosperms, and ferns [1–5]. The current concept of the genus was established in 2013, proposing the use of multilocus sequence analysis for species delimitation within the genus [4]. Since then, several protein-coding genes, such as actin (actA), β-tubulin (tub2), calmodulin (cmdA), glyceraldehyde-3phosphate dehydrogenase (gapdh), histone H3 (his3), translation elongation factor 1-alpha (tef1), and RNA polymerase II second largest subunit (rpb2), along with the internal transcribed spacer (ITS) region of the rDNA, have been investigated [6].
Among the poorly known and fastidious species of Cercospora and cercosporoid fungi, C. flagellaris Ellis & G. Martin remains unresolved, and its taxonomy has become increasingly complex with the emergence of new reports globally. This fungus was first described in 1882 as a leaf spot pathogen of Phytolacca decandra L. (currently Phytolacca americana L.) in North America, which is the native range of the host [7]. Numerous isolates that phylogenetically cluster within the flagellaris group have been designated as confer (cf.) flagellaris owing to several uncertainties. Bakshi et al. [6] noted that the cf. flagellaris group can be divided into three distinct clades based on analyses of eight gene loci in molecular phylogenetics. However, this remains insufficient for species delimitation because of overlaps in morphology and host ranges among these clades within the group. Currently, the number of C. cf. flagellaris isolates from various hosts continues to expand, encompassing plants from diverse families [4,8,9]. However, C. f lagellaris s. str. on Phytolacca spp. has been reported in North America and East Asia, with the exception of Ethiopia [10,11]. To date, only three isolates of C. flagellaris have been analyzed using multigene sequence data by Groenewald et al. [4], all of which originated from Korean samples of P. americana.
In Korea, this fungus was first reported on Phytolacca esculenta Van Houtte [12], followed by a report on P. americana [13]. The authors noted abundant hypophyllous fructification of the fungus on some samples that co-occurred with epiphyllous fructification. They compared their findings with Chupp’s description of American specimens, which were characterized by epiphyllous caespituli.
The first step in resolving the complexity of the flagellaris group was to elucidate the morphology and molecular phylogeny of true C. flagellaris on its original host. Therefore, this study aimed to reassess and integrate previous records with newly obtained collections of C. flagellaris on P. americana in Korea. By combining morphological examinations with multigene phylogenetic analyses, this study sought to clarify the identity of true C. flagellaris on its original host.
The samples used in this study are listed in Table 1, along with their collection dates, localities, strains, and sequence deposition IDs in the relevant databases. All collected samples were preserved at the Herbaria of Korea University (labeled as KUS-F, Seoul, Korea) and Jeonbuk National University (as JBNU-F, Jeonju, Korea). For morphological characterization of C. flagellaris, fungal structures were examined and photographed using a Zeiss AX10 microscope equipped with an AxioCam MRc5 camera (Carl Zeiss, Oberkochen, Germany). The size of each diagnostic feature was determined based on at least 20–30 measurements. To obtain single-spore isolates, conidia collected from young lesions were mounted on a drop of sterile water and streaked onto 2% water agar (WA; Junsei, Tokyo, Japan) plates supplemented with 100 mg/L streptomycin sulfate. The plates were incubated at 25°C. After 2 days, the germinating conidia were transferred to 2% potato dextrose agar (PDA; Difco, Detroit, MI, USA) plates to obtain a pure culture. Colony color was determined using Raynar color charts [14]. The obtained strains were deposited in the Korean Agricultural Culture Collection (KACC; Wanju, Korea).
All examined specimens were consistent with the leaf symptoms and fungal morphology. Initial lesions on the affected leaves were circular to irregular, brown to pale brown, and more or less sunken in the middle. As the spots enlarged with narrow borders, they became tan, creamy white, and even grayish with heavy fructification. Discrete lesions were mainly less than 6 mm in diameter, occasionally confluent, and cottony with numerous conidiophores and conidia (Fig. 1A, B). Stromata were small to medium, poorly developed, sometimes only occupying stomatal openings, brown to dark brown, 10–24 μm in diameter, and composed of several dark brown cells (Fig. 1C, D). Conidiophores were 8–16 in a divergent fascicle, emerging through stomatal openings in the early stage or from the cuticle of the upper leaf surface in the later stages of disease development. They were usually not branched, mildly or abruptly geniculated 1–4 times, olivaceous brown throughout or paler upwards, variable in length, 30–265 × 3–5 μm, and 0–5-septate (Fig. 1E). Conidial scars on conidiophores were conspicuous, apical, or at the position of geniculations, and 3–5 μm in diameter. Conidiogenesis was holoblastic, with terminal conidiogenous cells. Conidia were solitary, acicular-filiform, straight to mildly curved, hyaline, 4–12-septate, 40–160 × 3.0–4.5 μm, with conspicuously thickened and darkened hila, base truncate to subtruncate, and apex obtuse (Fig. 1F).

Fig. 1. Cercospora flagellaris s. str. on Phytolacca americana (micrographs were obtained from the sample JBNU-F0463). A: Infected plants showing leaf spots in the field; B: Young and mature leaf spots coalescing into necrotic lesions on the affected leaf; C: Close-up view of the leaf spot; D: Close-up view of fructification under a stereo microscope; E: Conidiophore; F: Conidia; G: One-week-old colonies grown on potato dextrose agar (PDA) at 25°C (obverse surface); H: Reverse surface of the same colony. Scale bars = 30 µm.
Two-week-old colonies grown on PDA at 25°C were 35–43 mm in diameter, with entire edge, the obverse surface was radially striate and cottony white in color, with a slightly pinkish margin. The reverse of the colonies was bloody red, and pinkish near the margin (Fig. 1G, H). The aforementioned morphological characteristics align with those of C. flagellaris reported by Chupp from North American samples [7] and Katsuki and Kobayashi from Japanese materials [15].
Genomic DNA was extracted from mycelia obtained from 2- or 3-week-old colonies using MaglistoTM 5M kits (Bioneer, Daejeon, Korea) according to the manufacturer’s guidelines. Nucleotide sequences of the ITS region and protein-coding genes such as actA, cmdA, tef1, and his3 were amplified and sequenced using ITS5/ITS4, ACT-512F/ACT-783R, CAL-228F/CAL-737R, EF1-728F/EF1-986R, and CylH3F/ CylH3R, respectively [16–18]. The PCR products were purified and sequenced using the same primers from Bioneer Inc. (Daejeon, Korea). The obtained forward and reverse sequences were inspected using BioEdit 7.2.5 [19], then assembled in MEGA 11 [20], and concatenated data were deposited in GenBank (Table 1).
Table 1. Information on Korean specimens of Cercospora flagellaris on Phytolacca americana used in this study
테이블
KACC: Korean Agricultural Culture Collection, Rural Development Administration; ITS: internal transcribed spacer; actA: actin; cmdA: calmodulin; his3: histone H3; tef1: translation elongation factor 1-alpha.
Multigene phylogenetic analyses were performed using a combined dataset of five genes. In total, 50 sequences of isolates were used, of which six were obtained in this study. Septoria provencialis (CBS 118910) was designated as an outgroup [9]. Information on the sequences used in the phylogenetic analyses is provided in Supplementary Table. Each gene was first aligned individually in MEGA 11 using the MUSCLE algorithm [20] and then concatenated into a single multilocus dataset of ITS+tef1+act+cmdA+his3 using SequenceMatrix software [21]. Character set partition was as follows: ITS 1–480, tef1 481–789, actA 790–1,003, cmdA 1,004–1,299, and his3 1,300–1,677 bp. The final data matrix consisted of 50 sequences and 1,677 characters. Character-based methods, maximum parsimony (MP), and maximum likelihood (ML) were used to generate phylogenetic trees in PAUP*4.0a using a heuristic search option and in raxmlGUI 2.0.14 using GTR+G [22,23]. Bootstrap analyses were performed with 1,000 replicates to evaluate the robustness of the internal branches [24].
The nucleotide sequences of ITS, actA, cmdA, his3, and tef1 were determined for six Cercospora strains isolated from P. americana in this study. We manually compared the genetic identities of these isolates with each other and with three previously reported Phytolacca-derived isolates (CPC 10684, CPC 10124, and CBS 132674). The ITS, cmdA, his3, and tef1 sequences were identical among all six strains, whereas the ITS and his3 sequences were identical to those of the three previously reported isolates. However, a single nucleotide polymorphism (SNP) was detected in the actA gene (226 bp), where adenine (A) was replaced by guanine (G) in KACC 43108 and 410484, distinguishing these two isolates from the remaining four and three isolates. A BLASTn search showed 100% identity to C. cf. flagellaris for ITS (KT193679), actA (DQ835121, JX143112, JX143123), cmdA (OQ773733, OQ773748), his3 (JX142625, JX142626, DQ835175), and tef1 (JX143360, JX143362).
The tree topologies generated by the MP and ML analyses were consistent. In total, 34 trees were retained from parsimony analysis. All nucleotide substitutions were equally weighted and unordered, and gaps were treated as missing data. Of the 1,677 total characters, 159 (9.4%) were variable and parsimonyuninformative, and 238 (14.1%) were informative for parsimony analysis. Tree scores were calculated, including tree length (TL = 774), consistency index (CI = 0.6589), retention index (RI = 0.7805), and rescaled consistency index (RC = 0.5143). Phylogenetic analysis revealed that all isolates obtained from P. americana, including six newly generated and three previously deposited sequences (CBS 132674, CPC 10124, and CPC 10684), formed a well-supported monophyletic subclade within the broader C. cf. flagellaris lineage (Fig. 2). This subclade was strongly supported (79/82% BS), indicating that it was a genetically coherent group. In contrast, other isolates identified as C. cf. flagellaris, which were derived from diverse host plants, such as Amaranthus hybridus, Cichorium intybus, Sigesbeckia pubescens, were distributed across several more heterogeneous and weakly supported branches. This branching pattern suggests that C. cf. flagellaris is a species complex with considerable host-associated divergences. The consistent clustering of Phytolacca-derived isolates into distinct and well-supported lineages supports the hypothesis that these isolates represent C. flagellaris sensu stricto (s. str.).
P. americana (Phytolaccaceae), commonly known as the American pokeweed, is native to North America. It is now widely distributed in North America, South America, Africa, and several countries in Asia [25]. This plant was introduced to Korea in the 1950s and is currently distributed across various locations on the peninsula, including areas near factories and vacant lots [26]. To date, C. flagellaris s. str. is native to North America and has become invasive in Japan, Korea [12,13,15,27], and Taiwan [28]. Phylogenetic analysis based on multiple genes is required for this fungus from American samples to prove the true identity of C. flagellaris. As this species was originally described as a leaf spot fungus on P. americana, the concept of C. flagellaris should be limited to isolates obtained from Phytolacca species. Therefore, we propose that C. flagellaris s. str. as a standard for the isolates. This is the first comprehensive documentation of C. flagellaris on P. americana in Korea.
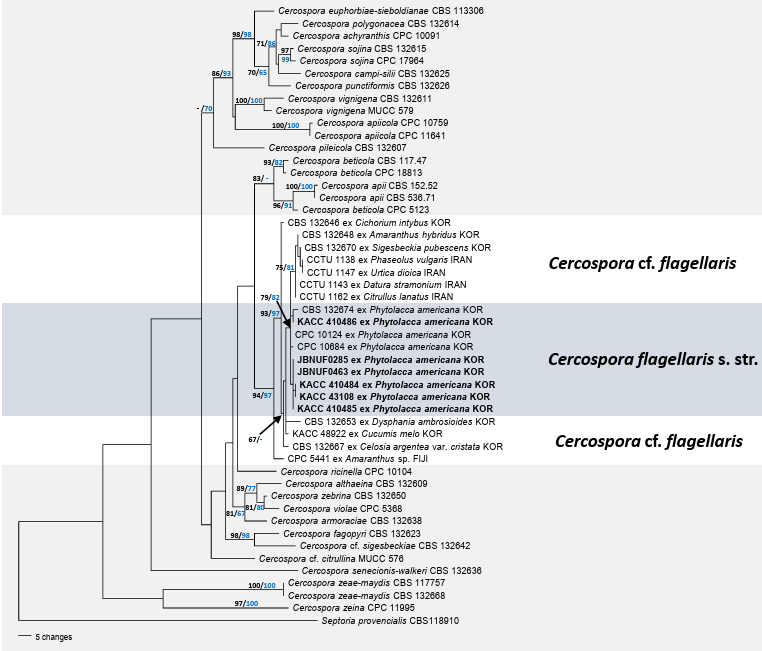
Fig. 2. Parsimonious tree of Cercospora highlighting C. flagellaris. The tree was generated from a combined multigene dataset of ITS + tef1 + actA + cmdA + his3, comprising 50 sequences and 1,677 characters. All isolates obtained in this study are indicated in bold. The branching patterns and topologies of the trees from the maximum parsimony (MP) and maximum likelihood (ML) analyses were consistent. Bootstrap values (>70%) obtained using MP (black) and ML (blue) are displayed on the branch. ITS: internal transcribed spacer; actA: actin; cmdA: calmodulin; his3: histone H3; tef1: translation elongation factor 1-alpha.
No potential conflict of interest was reported by the author(s).
This research was supported by the Global-Learning & Academic Research Institute for Master’s PhD students and Postdocs (LAMP) Program of the National Research Foundation of Korea (NRF) grant funded by the Ministry of Education (No. RS-2024-00443714).
1. Braun U, Nakashima C, Crous PW. Cercosporoid fungi (Mycosphaerellaceae) 1. Species on other fungi, Pteridophyta and Gymnospermae. IMA Fungus 2013;4:265-345. https://doi. org/10.5598/imafungus.2013.04.02.12 [DOI]
2. Braun U, Crous PW, Nakashima C. Cercosporoid fungi (Mycosphaerellaceae) 2. Species on monocots (Acoraceae to Xyridaceae, excluding Poaceae). IMA Fungus 2014;5:203-390. https://doi.org/10.5598/imafungus.2014.05.02.04 [DOI]
3. Braun U, Crous PW, Nakashima C. Cercosporoid fungi (Mycosphaerellaceae) 3. Species on monocots (Poaceae, true grasses). IMA Fungus 2015;6:25-97. https://doi.org/10.5598/ imafungus.2015.06.01.03 [DOI]
4. Groenewald JZ, Nakashima C, Nishikawa J, Shin HD, Park JH, Jama AN, Groenewald M, Braun U, Crous PW. Species concepts in Cercospora: Spotting the weeds among the roses. Stud Mycol 2013;75:115-70. https://doi.org/10.3114/sim0012 [DOI]
5. Guatimosim E, Schwartsburd PB, Barreto RW, Crous PW. Novel fungi from an ancient niche: Cercosporoid and related sexual morphs on ferns. Persoonia 2016;37:106-41. https://doi. org/10.3767/003158516×690934 [DOI]
6. Bakhshi M, Arzanlou M, Babai-Ahari A, Groenewald JZ, Crous PW. Novel primers improve species delimitation in Cercospora. IMA Fungus 2018;9:299-332. https://doi.org/10.5598/ imafungus.2018.09.02.06 [DOI]
7. Chupp CA. A monograph of the fungus genus Cercospora. Ithaca, NY: Cornell University Press; 1954.
8. Bakhshi M, Arzanlou M, Babai-Ahari A, Groenewald JZ, Braun U, Crous PW. Application of the consolidated species concept to Cercospora spp. from Iran. Persoonia 2015;34:65-86. https://doi.org/10.3767/003158515×685698 [DOI]
9. Park MJ, Back CG, Park JH. Occurrence of Cercospora leaf spot caused by Cercospora cf. flagellaris on melon in Korea. Mycobiology 2020;48:418-22. https://doi.org/10.1080/1229 8093.2020.1792133 [DOI]
10. Crous PW, Braun U. Mycosphaerella and its anamorphs: 1. Names published in Cercospora and Passalora. Utrecht, Netherlands: Centraalbureau voor Schimmelcultures; 2003.
11. Farr DF, Rossman AY. Fungal databases, US national fungus collections, Agricultural Research Service (ARS), United States Department of Agriculture (USDA) [Internet]. Beltsville, MD: USDA; 2025 [cited 2025 Jun 17]. Available from: https://fungi.ars.usda.gov/.
12. Kim JD, Shin HD. Taxonomic studies on Cercospora and allied genera in Korea (VIII). Kor J Mycol 1999;27:147-57.
13. Shin HD, Kim JD. Cercospora and allied genera from Korea. Suwon, Korea: National Institute of Agricultural Science and Technology; 2001.
14. Raynar RW. A mycological color chart. Kew, UK: Commonwealth Mycological Institute and British Mycological Society; 1970.
15. Katsuki S, Kobayashi T. Cercosporae of Japan and allied genera (supplement 5). Trans Mycol Soc Jpn 1982;23:41-9.
16. White TJ, Bruns TD, Lee SB, Taylor JW. Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics. In: Innis MA, Gelfand DH, Sninsky JJ, editors. PCR protocols: A guide to methods and applications. New York: Academic Press; 1990. p. 315-22. [DOI]
17. Carbone I, Kohn LM. A method for designing primer sets for speciation studies in filamentous ascomycetes. Mycologia 1999;91:553-6. https://doi.org/10.1080/00275514.1999.12061051 [DOI]
18. Crous PW, Groenewald JZ, Risède JM, Simoneau P, Hywel-Jones NL. Calonectria species and their Cylindrocladium anamorphs: species with sphaeropedunculate vesicles. Stud Mycol 2004;50:415-30.
19. Hall TA. BioEdit: A user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucleic Acids Symp Ser 1999;41:95-8.
20. Tamura K, Stecher G, Kumar S. MEGA11: Molecular evolutionary genetics analysis version 11. Mol Biol Evol 2021;38:3022-7. https://doi.org/10.1093/molbev/msab120 [DOI]
21. Vaidya G, Lohman DJ, Meier R. SequenceMatrix: Concatenation software for the fast assembly of multi-gene datasets with character set and codon information. Cladistics 2011;27:171-80. https://doi.org/10.1111/j.1096-0031.2010.00329.x [DOI]
22. Swofford DL. PAUP: Phylogenetic analyses using parsimony (*and Other Methods) 4.0a10. Sunderland, MA: Sinauer Associates. 2002.
23. Silvestro D, Michalak I. RaxmlGUI: A graphical front-end for RAxML. Org Divers Evol 2012;12:335-7. https://doi.org/10.1007/s13127-011-0056-0 [DOI]
24. Felsenstein J. Confidence limits on phylogenies: An approach using the bootstrap. Evolution 1985;39:783-91. https://doi.org/10.1111/j.1558-5646.1985.tb00420.x [DOI]
25. Follak S, Schwarz M, Essl F. Notes on the occurrence of Phytolacca americana L. in crop fields and its potential agricultural impact. Bioinvasions Rec 2022;11:620-30. https://doi. org/10.3391/bir.2022.11.3.04 [DOI]
26. Min BM. Distribution of Phytolacca americana in a coastal sand dune. J Ecol Environ 2014;37:81-90. https://doi.org/10.5141/ecoenv.2014.010 [DOI]
27. Shin HD, Braun U. Notes on Korean Cercosporae and allied genera (III). Mycotaxon 2000;74:105-18. [DOI]
28. Kirschner R. A new species and new records of cercosporoid fungi from ornamental plants in Taiwan. Mycol Prog 2014;13:483-91. https://doi.org/10.1007/s11557-013-0930-6 [DOI]
Supplementary table. Information on sequences derived from various Cercospora isolates used in the phylogenetic analyses.
테이블
ITS: internal transcribed spacer; actA: actin; cmdA: calmodulin; his3: histone H3; tef1: translation elongation factor 1-alpha.