Yeon-Su Jeong1, Seong-Keun Lim1, Geun-A Park1, Seung-Yeol Lee1,2, and Hee-Young Jung1,2*
1Department of Plant Medicine, Kyungpook National University, Daegu 41566, Korea
2Institute of Plant Medicine, Kyungpook National University, Daegu 41566, Korea
*Correspondence to heeyoung@knu.ac.kr
Korean Journal of Mycology (Kor J Mycol) 2025 September, Volume 53, Issue 3, pages 203-210.
https://doi.org/10.4489/kjm.2025.53.3.7
Received on August 14, 2025, Revised on September 11, 2025, Accepted on September 11, 2025, Published on September 30, 2025.
Copyright © The Korean Society of Mycology.
This is an Open Access article which is freely available under the Creative Commons Attribution-Non-Commercial 4.0 International License (CC BY-NC) (https://creativecommons.org/licenses/by-nc/4.0/).
As part of a fungal taxonomic study in Korea, strain KNU-YC-1803F was isolated from soil collected in Gogyeong-myeon, Yeongcheon-si, Gyeongsangbuk-do. When cultivated on potato dextrose agar and Sabouraud dextrose agar, the colonies appeared fluffy white with a pale yellow reverse. Microscopic examination revealed the presence of aleurioconidia that were hyaline, clavate to elongated-pyriform, measuring approximately 9.2 × 6.1 μm. Additionally, arthroconidia were abundantly formed in chains, cylindrical to navicular in shape, and measured around 7.6 × 5.7 μm. Sequence analysis of the internal transcribed spacer (ITS) region showed 100% similarity with Trichophyton verrucosum and 99.8% similarity with Chrysosporium articulatum and Aphanoascus reticulisporus. Complementary analysis of the large subunit (LSU) of the nuclear ribosomal DNA revealed high similarity to several related taxa. When trimmed for phylogenetic analysis, the LSU sequence exhibited 100% similarity to multiple species, including C. articulatum. The phylogenetic tree, constructed from concatenated ITS and LSU sequences, revealed that the isolate clustered within a clade containing C. articulatum, distinctly separated from other closely related taxa. Taken together, the morphological characteristics and molecular phylogenetic evidence confirm that strain KNU-YC-1803F is C. articulatum, marking the first documented occurrence of this species in Korea.
Chrysosporium articulatum, Molecular phylogeny, Morphological characterization, Phylogenetic analysis, Soil-derived fungi
The family Onygenaceae (Ascomycota) is characterized by hyaline mycelia and the formation of arthrospores and aleuriospores, typically producing hyaline conidia. Members of this family are frequently isolated from soil, plant debris, and keratin-rich substrates and include genera such as Chrysosporium, Aphanoascus, and Onygena [1].
The genus Chrysosporium, first described by Corda in 1833 [2], currently encompasses approximately 100 recognized species worldwide [3]. Species of this genus typically form white to pale colonies and produce simple conidia that are subglobose, pyriform, or clavate in shape [4]. However, morphological similarities among Onygenaceae species often complicate accurate identification. To address this issue, molecular phylogenetic analyses-particularly those based on internal transcribed spacer (ITS) sequences-are now widely employed alongside traditional morphological observations [5]. Consequently, several species originally classified under Chrysosporium have been reallocated to related genera such as Aphanoascus or Keratinophyton [6].
Chrysosporium species are widely distributed across diverse environments, including terrestrial soils, animal-associated habitats, and marine ecosystems [7]. They are often recovered from keratin-rich materials such as feathers, hair, and skin, reflecting their ability to secrete keratin-degrading enzymes and other secondary metabolites [3,8,9]. These properties facilitate the degradation of keratinous waste, promote nutrient recycling in ecosystems, and offer considerable potential for industrial applications. In Korea, several Chrysosporium species have recently been reported, most of which were isolated from soil [4,10], while others were recovered from freshwater sediment [11].
During a survey of fungal diversity in Korean soils, strain KNU-YC-1803F was isolated. This study presents the first record of this species in Korea and aims to determine its precise taxonomic placement. To achieve reliable identification, detailed morphological characterization and molecular phylogenetic analyses were conducted.
The fungal strain described in this study was isolated from a soil sample collected in Gogyeong-myeon, Yeongcheon-si, Gyeongsangbuk-do, Republic of Korea (35°57′56.39′′N, 128°59′27.53′′E). Isolation was performed using the serial dilution method. A 100 μL aliquot of the soil suspension was spread onto potato dextrose agar (PDA; Difco, Detroit, MI, USA) and incubated at 25°C under dark conditions for two weeks [12]. Subsequently, distinct single colonies were selected and transferred to fresh PDA plates for purification. Preliminary identification was conducted based on morphological characteristics, genomic DNA extraction, PCR amplification, and sequencing of the ITS regions. The isolate, designated KNU-YC-1803F, was tentatively identified as an unrecorded species in Korea. Strain KNU-YC-1803F (NIBRFGC000502236) has been deposited in a metabolically inactive state at the National Institute of Biological Resources (NIBR), Republic of Korea.
Strain KNU-YC-1803F was cultured on PDA and Sabouraud dextrose agar (SDA; MBcell, Seoul, Korea) at 25°C under dark conditions. To observe colony morphology, the strain was incubated for four weeks. Colony features, including color, texture, and growth pattern, were documented using a Canon EOS 5D Mark III digital camera (Canon, Tokyo, Japan). To observe microscopic morphological features such as spore shape and size, the strain was cultured on a glass slide for 1–2 weeks and examined using a light microscope (BX-50, Olympus, Tokyo, Japan).
Genomic DNA of strain KNU-YC-1803F was extracted using the HiGene Genomic DNA Prep Kit (BIOFACT, Daejeon, South Korea), following the manufacturer’s instructions. The ITS regions were amplified using primers ITS1F and ITS4 [13], while the large subunit (LSU) of the nuclear ribosomal RNA gene was amplified using the primers LR0R and LR7 [14]. PCR products were purified using the ExoSAPIT reagent (Thermo Fisher Scientific, Waltham, MA, USA) and sequenced by Macrogen (Seoul, South Korea). Sequences of the ITS region and LSU gene were deposited in GenBank under accession numbers LC880189 and LC880190, respectively.
For molecular phylogenetic analysis, the sequences of strain KNU-YC-1803F were compared with reference sequences available in the GenBank database of the National Center for Biotechnology Information (NCBI) using the Basic Local Alignment Search Tool (BLAST). Closely related sequences were retrieved from GenBank for further analysis (Table 1). Multiple sequence alignment was performed using ClustalX version 2.0 (European Bioinformatics Institute, Hinxton, United Kingdom). Phylogenetic analyses were conducted using MEGA version 7.0 (Institute of Molecular Evolutionary Genetics, Pennsylvania State University, University Park, PA, USA) [15]. A concatenated dataset of ITS and LSU sequences was used to construct the phylogenetic tree. The tree was inferred using the maximum likelihood (ML) method with 1,000 bootstrap replicates. Genetic distances between taxa were calculated using Kimura’s two-parameter model [16].
Table 1. GenBank accession numbers of sequences used for phylogenetic analyses in this study
테이블
Strain KNU-YC-1803F formed colonies measuring 21.6–23.2 mm (mean = 22.4 ± 0.6 mm, n = 6) in diameter after 7 days and 41.7–45.4 mm (mean = 44.2 ± 1.3 mm, n = 6) after 14 days of incubation on PDA. On SDA medium, the colony diameter reached 26.1–26.8 mm (mean = 26.6 ± 0.4 mm, n = 4) after 7 days and 40.8–43.6 mm (mean = 42.0 ± 1.4 mm, n = 4) after 10 days of incubation. Colonies on both PDA and SDA were white and fluffy in appearance, with a pale yellow coloration on the reverse side (Fig. 1 A–D).
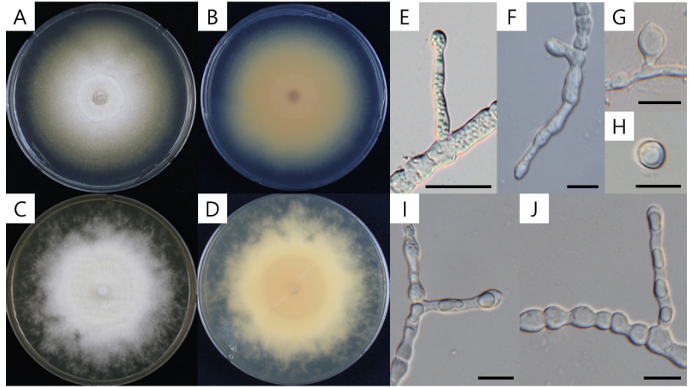
Fig. 1. Culture and morphological characteristics of Chrysosporium articulatum. A: Surface of colony on potato dextrose agar (PDA); B: Reverse of colony on PDA; C: Surface of colony on Sabouraud dextrose agar (SDA); D: Reverse of colony on SDA; E–H: Aleurioconidia; I and J: Arthroconidia. Scale bars: E, 20 µm; F–J, 10 µm.
Microscopic examination revealed aleurioconidia that were hyaline, clavate, and elongated-pyriform in shape, measuring 4.1–14.0 × 4.4–9.2 μm (mean = 9.2 ± 2.4 × 6.1 ± 1.2 μm, n = 30) (Fig. 1 EH). Arthroconidia were abundantly produced, cylindrical to navicular in shape, and formed in chains, with dimensions of 4.7–11.9 × 3.6–7.7 μm (mean = 7.6 ± 1.7 × 5.7 ± 0.9 μm, n = 60) (Fig. 1 I & J). The colony morphology and microscopic features of KNU-YC-1803F closely resembled those of C. articulatum [8,17] (Table 2). In contrast, C. keratinophilum forms fluffy yellow colonies with a cream to brown reverse, and its aleurioconidia are larger (9–13 × 6–9 μm) than those of KNU-YC-1803F [18]. Additionally, arthroconidia are rarely observed in C. keratinophilum. Based on these morphological distinctions, KNU-YC-1803F is most similar to C. articulatum and can be clearly differentiated from C. keratinophilum.
Table 2. Comparison of morphological and cultural characteristics between KNU-YC-1803F and the phylogenetically closest species of the genus Chrysosporium
테이블
The ITS sequence of KNU-YC-1803F was determined to be 618 bp in length. BLAST analysis using the NCBI database revealed 100% similarity with Trichophyton verrucosum (NIAS CH101; ON843702) and 99.8% similarity with Chrysosporium articulatum (UAMH 4320; AJ007841) and Aphanoascus reticulisporus (IHEM:23961; OW987155). The ITS match with T. verrucosum likely reflects limitations of the sequences available in the NCBI database, rather than indicating a close phylogenetic relationship. Although recent studies have relied solely on ITS sequences for species delimitation within the genus Chrysosporium [5], the present study indicates that ITS alone may be insufficient for reliable genus- and species-level identification. Therefore, LSU sequencing was performed to enable multi-locus phylogenetic analysis.
The LSU sequence of KNU-YC-1803F was determined to be 1,317 bp in length. BLAST analysis of the LSU sequence showed 99.9% similarity to Aphanoascus guizhouensis (ZY 22.044; OR680590) and 96.8% similarity to Keratinophyton sichuanense (CGMCC 3.20871; NG_149049) and Keratinophyton hubeiense (CGMCC 3.20870; OM952116). After trimming for phylogenetic analysis, the LSU sequence showed 100% similarity to A. reticulisporus (NBRC 32372; JN941549), Chrysosporium keratinophilum (CBS 392.67; AY176730.1), and C. articulatum (FMR 19012; ON720740).
To determine the phylogenetic placement of strain KNU-YC-1803F, a phylogenetic tree was constructed from concatenated ITS and LSU sequences using the ML method. The results showed that KNU-YC1803F clustered with C. articulatum and was clearly separated from C. keratinophilum, supporting its identification as C. articulatum (Fig. 2).
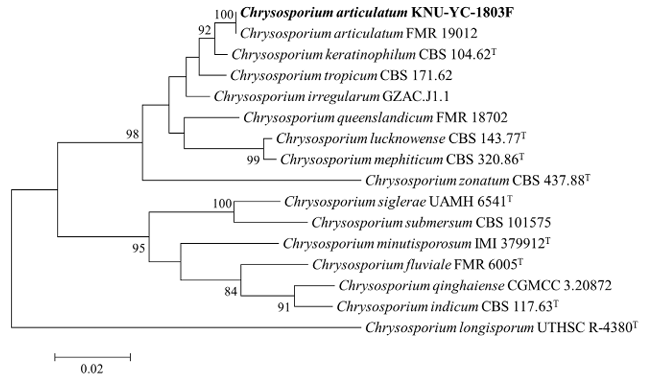
Fig. 2. Maximum likelihood phylogenetic tree based on internal transcribed spacer (ITS) regions and large subunit (LSU) gene sequences showing the phylogenetic position of KNU-YC-1803F within the genus Chrysosporium. Chrysosporium longisporum UTHSC R-4380T was used as the outgroup. The isolate from this study is highlighted in bold. The numbers above the branches indicate bootstrap values (>80%) from 1,000 replicates. Bar = 0.02 substitutions per nucleotide position.
Previously, C. articulatum and Chrysosporium queenslandicum were considered conspecific, based on their presumed connection to the teleomorph Apinisia queenslandica [2]. However, this classification relied primarily on morphological similarity and lacked molecular phylogenetic evidence, rendering it a limited and provisional interpretation. Later, Sigler reclassified the two species as distinct based on clear morphological differences, including the arrangement of arthroconidia and the size and structural characteristics of conidia [19]. More recently, molecular phylogenetic analysis based on ITS sequences further supported this distinction, demonstrating that the two species occupy separate clades consistent with their morphological divergence [5]. Furthermore, phylogenetic analyses based on concatenated ITS and LSU sequences placed KNU-YC-1803F within the same clade as C. articulatum, clearly separating it from C. queenslandicum and supporting its identification as C. articulatum (Fig. 2).
Chrysosporium articulatum was previously reported from an unidentified marine sponge in Korea [20]; however, that study lacked definitive morphological, molecular, and cultural evidence, rendering the identification uncertain. In the present study, the species was confirmed as C. articulatum through detailed morphological observation and multi-locus phylogenetic analysis based on ITS and LSU sequences. Accordingly, this strain is herein reported as the first confirmed record of C. articulatum in Korea, carrying significant taxonomic value.
The genus Chrysosporium is known to produce a wide array of secondary metabolites, with approximately 66% identified as novel natural compounds. These include alkaloids, polyketides, lactones, and furan derivatives, many of which exhibit notable bioactivities such as cytotoxicity, antibacterial, antifungal, and enzyme inhibitory effects [7]. In particular, C. articulatum and several related species have been reported to produce keratinase, an enzyme capable of degrading animal keratin such as feathers and hair [8], suggesting its potential application in keratin waste management and industrial enzyme production.
The domestic report of this strain extends beyond mere identification, representing a meaningful contribution to the expansion of national fungal biodiversity from soil environments and serving as a foundational resource for strain banking and future biotechnological utilization.
The authors declare that they have no potential conflicts of interest.
This research was supported by the National Institute of Biological Resources, funded by the Ministry of Environment of the Republic of Korea [NIBR201801105].
1. Malloch D, Cain RF. New genera of Onygenaceae. Can J Bot 1971;49:839-46. [DOI]
2. van Oorschot CAN. A revision of Chrysosporium and allied genera. Stud Mycol 1980;20:1-89.
3. Kizerwetter-Świda M, Bąk I, Biegańska MJ, Dembele K, Chrobak-Chmiel D. Chrysosporium articulatum mimicking Trichophyton spp. infection in a cat: A case presentation and literature review. BMC Vet Res 2024;20:359. https://doi.org/10.1186/s12917-024-04185-7 [DOI]
4. Gurung SK, Adhikari M, Kim SW, Bazie S, Kim HS, Lee HG, Kosol S, Lee HB, Lee YS. Discovery of two Chrysosporium species with keratinolytic activity from field soil in Korea. Mycobiology 2018;46:260-8. https://doi.org/10.1080/12298093.2018.1514732 [DOI]
5. Han YF, Ge W, Zhang ZY, Liang JD, Chen WH, Huang JZ, Liang ZQ. Morphological and phylogenetic characterisations reveal nine new species of Chrysosporium (Onygenaceae, Onygenales) in China. Phytotaxa 2022;539:1-16. https://doi.org/10.11646/phytotaxa.539.1.1 [DOI]
6. Kandemir H, Dukik K, de Melo Teixeira M, Stielow JB, Delma FZ, Al-Hatmi AMS, Ahmed SA, Ilkit M, de Hoog GS. Phylogenetic and ecological reevaluation of the order Onygenales. Fungal Divers 2022;115:1-72. https://doi.org/10.1007/s13225-022-00506-z [DOI]
7. Wang Y, Yang X, Li Y, Wang B, Shi T. The genus Chrysosporium: A potential producer of natural products. Fermentation 2023;9:76. https://doi.org/10.3390/fermentation9010076 [DOI]
8. Bohacz J. Biodegradation of feather waste keratin by a keratinolytic soil fungus of the genus Chrysosporium and statistical optimization of feather mass loss. World J Microbiol Biotechnol 2017;33:13. https://doi.org/10.1007/s11274-016-2177-2 [DOI]
9. English MP. Destruction of hair by Chrysosporium keratinophilum. Trans Br Mycol Soc 1969;52:247-55. https://doi.org/10.1016/S0007-1536(69)80037-9 [DOI]
10. Phaund W, Somaly U, Das K, Lee SY, Jung HY. Clonostachys divergens and Chrysosporium merdarium: Two new records from soil in Korea. Kor J Mycol 2023;51:91-100. [DOI]
11. Goh J, Mun HY, Oh Y. Seven previously unrecorded fungal species isolated from freshwater ecosystems in Korea. Kor J Mycol 2021;49:183-97. [DOI]
12. Jeong YS, Lim SK, Nam SW, Ten LN, Lee SY, Jung HY. Scytalidium terrigenum sp. nov., a new species isolated from soil in Korea. Mycobiology 2025;53:295-304. https://doi.org/10.10 80/12298093.2025.2479241 [DOI]
13. White TJ, Bruns TD, Lee SB, Taylor JW. Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics. In: Innis MA, Gelfand DH, Sninsky JJ, editors. PCR protocols: A guide to methods and applications. New York: Academic Press; 1990. p. 315-22. [DOI]
14. Rehner SA, Samuels GJ. Taxonomy and phylogeny of Gliocladium analysed from nuclear large subunit ribosomal DNA sequences. Mycol Res 1994;98:625-34. [DOI]
15. Kumar S, Stecher G, Tamura K. MEGA7: Molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol Biol Evol 2016;33:1870-4. https://doi.org/10.1093/molbev/ msw054 [DOI]
16. Kimura M. A simple method for estimating evolutionary rates of base substitutions through comparative studies of nucleotide sequences. J Mol Evol 1980;16:111-20. https://doi. org/10.1007/bf01731581 [DOI]
17. Scharapov VM. Species Chrysosporii Corda novae. Nov Syst Niz Rast 1978;15:141-9.
18. Otčenášek M, Dvořák J. The isolation of Chrysosporium keratinophilum (Frey) Carmichael 1962 and similar fungi from Czechoslovakian soil. Mycopathol Mycol Appl 1964;23:121-4. https://doi.org/10.1007/BF02049267 [DOI]
19. Sigler L, Guarro J, Punsola L. New keratinophilic species of Chrysosporium. Can J Bot 1986;64:1212-5. https://doi.org/10.1139/b86-164 [DOI]
20. Jeon JE, Julianti E, Oh H, Park W, Oh DC, Oh KB, Shin J. Stereochemistry of hydroxybearing benzolactones: Isolation and structural determination of chrysoarticulins A-C from a marine-derived fungus Chrysosporium articulatum. Tetrahedron Lett 2013;54:3111-5. https:// doi.org/10.1016/j.tetlet.2013.04.006 [DOI]