Ju-Heon Lee1,2, Youngsoo Kim1, Jong-Taek Park1, Dong-Hyuk Lee1, and Hee-Young Jung2*
1Apple Research Center, National Institute of Horticultural & Herbal Science, Gunwi 43100, Korea
2Department of Plant Medicine, Kyungpook National University, Daegu 41566, Korea
*Correspondence to heeyoung@knu.ac.kr
Korean Journal of Mycology (Kor J Mycol) 2025 September, Volume 53, Issue 3, pages 211-219.
https://doi.org/10.4489/kjm.2025.53.3.8
Received on July 03, 2025, Revised on September 12, 2025, Accepted on September 15, 2025, Published on September 30, 2025.
Copyright © The Korean Society of Mycology.
This is an Open Access article which is freely available under the Creative Commons Attribution-Non-Commercial 4.0 International License (CC BY-NC) (https://creativecommons.org/licenses/by-nc/4.0/).
In the present study, we isolated a fungus from an ambrosia beetle collected from a trap in an apple orchard in Gunwi-gun, Daegu, South Korea. The isolate was designated as ARI-25-A11. To classify the strain at a species level, both morphological and molecular analyses were conducted, and its phylogenetic position was determined. For cultural characterization, ARI-25-A11 was cultured on 2% malt extract agar at 25°C for 10 days, resulting in a colony diameter of 30.4–32.9 mm. White mycelium was observed at the center, and as the incubation period progressed, the center gradually darkened from olive to brown. Morphologically, the conidiophores were mononematous, septate, and hyaline, whereas the conidia were aseptate, predominantly oval, with an enlarged upper part and a tapered base, measuring an average of 5.9 × 3.5 µm. For molecular identification, the large subunit ribosomal RNA (LSU), small subunit ribosomal RNA (SSU), and β-tubulin (β-tub) genes were amplified and sequenced. The LSU and SSU sequences showed 100% identity with those of Raffaelea promiscua PL1001, and the β-tub gene sequence completely matched that of both R. promiscua PL1001 and CMW55899T. Phylogenetic analysis identified ARI-25-A11 within the same clade as R. promiscua. Based on morphological and molecular data, ARI-25-A11 was identified as R. promiscua, representing the first report of this species among ambrosia beetles in Korea.
Ambrosia beetles, Raffaelea promiscua, Symbiotic fungi, Taxonomy
Ambrosia beetles are small wood-boring insects belonging to the family Curculionidae, with approximately 3,500 species reported till date. They are classified into two subfamilies, Platypodinae and Scolytinae [1]. Most beetles maintain obligate mutualistic relationships with fungi, with female beetles primarily collecting fungal symbionts and storing them in specialized structures called mycangia, enabling the transmission of symbionts across generations [2]. Most species of ambrosia beetles harbor fungal symbionts belonging to the orders Ophiostomatales and Microascales, both within Ascomycota [3]. Among these, the genera Afroraffaelea [4], Aureovirgo [5], and Raffaelea [6], within Ophiostomatales, are considered the major fungal symbionts of ambrosia beetles.
The genus Raffaelea, belonging to the phylum Ascomycota and family Ophiostomataceae, was established by Arx & Hennebert [7] to accommodate Raffaelea ambrosiae isolated from beetles of the genus Platypus. Although most Raffaelea species are saprotrophic, some exhibit strong pathogenicity and cause serious damage to forests and agriculture. Notable examples include R. quercivora, the causative agent of Japanese oak wilt [8]; R. quercus-mongolicae, reported in Korea [9]; and R. lauricola, which is responsible for laurel wilt in the United States [10].
In the past, owing to the limitations of taxonomic techniques, the distinction between Raffaelea and Ambrosiella was unclear. These genera are distinguished primarily by their conidiogenous cell development. In Raffaelea, conidiogenesis occurs through sympodial branching, in which new cells are produced laterally. In contrast, Ambrosiella exhibits percurrent proliferation in which new cells are formed through the elongation of existing conidiogenous cells. However, these morphological differences are subtle and difficult to discern under a microscope, often leading to frequent misclassifications of the two genera [11]. Recently, molecular phylogenetic approaches have been used to clarify the taxonomic relationships among these fungi. Ribosomal DNA sequence analysis revealed that Raffaelea belongs to the order Ophiostomatales, whereas Ambrosiella belongs to the order Microascales, confirming that these two genera are not closely related phylogenetically [12]. Accordingly, the aim of the present study was to clarify the taxonomic identity of fungal isolates obtained from ambrosia beetles collected from apple orchards, using morphological characterization and rDNA-based molecular phylogenetic analyses.
Ambrosia beetles were trapped in an apple orchard located at the Apple Research Center in Gunwi-gun, Daegu-si, Republic of Korea (36°29′68.9″N, 128°46′56.1″E). After collection, the beetles were surfacesterilized using 70% ethanol and allowed to dry in the air for approximately 10 min. Sterilized specimens were transferred onto potato dextrose agar (PDA; Difco, Detroit, MI, USA) and incubated at 25°C for 3 days. The fungal growth was transferred to fresh PDA plates and incubated again under the same conditions for 10 days to obtain pure cultures. One isolate, designated ARI-25-A11, was preserved in 20% glycerol at −80°C for long-term storage. The specimen was deposited in the Korean Agricultural Culture Collection (KACC) and assigned the accession number KACC 411081.
The isolate ARI-25-A11 was cultured on PDA and 2% malt extract agar (MEA; Oxoid, Basingstoke, UK) at 25°C for 10 days to assess its cultural and morphological characteristics. After incubation, various colony traits, such as the diameter and pigmentation, were examined. The structure and dimensions of the conidiophores and conidia were observed and documented using a light microscope (CX-43; Olympus, Japan).
Genomic DNA was extracted from ARI-25-A11 to compare its sequence with those of related strains in the National Center for Biotechnology Information (NCBI) database and determine its phylogenetic position. Partial sequences of the large subunit ribosomal RNA (LSU), small subunit ribosomal RNA (SSU), and β-tubulin (β-tub) genes were obtained through PCR amplification. The LSU genes were amplified using the LR0R/LR5 primer pair [13], the SSU genes were amplified using the NS1/NS4 primers [14], and the β-tub genes were amplified using the Bt2a/Bt2b primers [15].
The PCR amplicons were confirmed via electrophoresis on 1% agarose gels and visualized through ethidium bromide staining. The amplified DNA was purified using EXOSAP-IT (Thermo Fisher Scientific, Waltham, MA, USA) according to the manufacturer’s protocol. Sequencing was performed by Solgent Co., Ltd. (Daejeon, Korea), and the resulting reads were assembled and analyzed using the SeqMan module within the Lasergene software suite (DNAStar Inc., Madison, WI, USA). The finalized sequences were deposited in GenBank under the accession numbers LC878894 (LSU), LC878895 (SSU), and LC878896 (β-tub).
Nucleotide sequences of Raffaelea species were retrieved from the NCBI database (Table 1). Sequence alignment was performed using Clustal X 2.0, within the MEGA 7 software suite [16]. Concatenated LSU, SSU, and β-tub gene sequences were used to construct a phylogenetic tree. Phylogenetic analysis was conducted using the nearest neighbor interchange algorithm under Kimura’s two-parameter substitution model [17], with gap sites excluded from the dataset. The tree was generated using the maximum likelihood method [18], and node support was evaluated based on 1,000 bootstrap replicates.
Table 1. The following is a list of species included in the phylogenetic analyses, along with their corresponding GenBank accession numbers
테이블
LSU: large subunit ribosomal RNA; SSU: small subunit ribosomal RNA; β-tub: β-tubulin; N/A: Not available.
Tex-type.
The isolated strain is shown in bold.
When cultured on PDA at 25°C for 10 days, the colony diameter reached 31.5–33.5 mm (av. 32.4 mm). On the front side, the center appeared dark olive in color, with the color gradually fading toward the edges to appear white. Initially, the mycelia grew into the medium; however, over time, a small amount of aerial mycelia was also observed. The reverse side of the colony was entirely white, with a dark brown band (Fig. 1A, B). Under the same conditions on 2% MEA, the fungus exhibited slower growth, with colony diameters reaching 30.4–32.9 mm (av. 31.6 mm). The colonies were flat and circular, initially appearing white, but gradually turned olive and then dark brown from the center as incubation progressed. Most of the hyphae were submerged in the medium (Fig. 1C, D).
The conidiophores were mononematous, septate, and hyaline. They were mostly unbranched but occasionally showed branching (Fig. 1E). Typically upright and straight, the structures become narrower toward the apex and were sometimes simplified as conidiogenous cells. Conidiogenous cells were integrated, hyaline, and exhibited a blastic mode of development. They were cylindrical or peg-like in shape and narrow toward the apex (Fig. 1F, G). The conidia were hyaline, aseptate, and formed blastically. Moreover, they were round, oval, or oblong with an enlarged upper portion and a tapered base. Conidia were often clustered near the conidiogenous cells (Fig. 1H). Further, their size range was 5.2–6.6 × 2.8–4.2 µm (av. 5.9 × 3.5 µm, n = 50) (Fig. 1I).
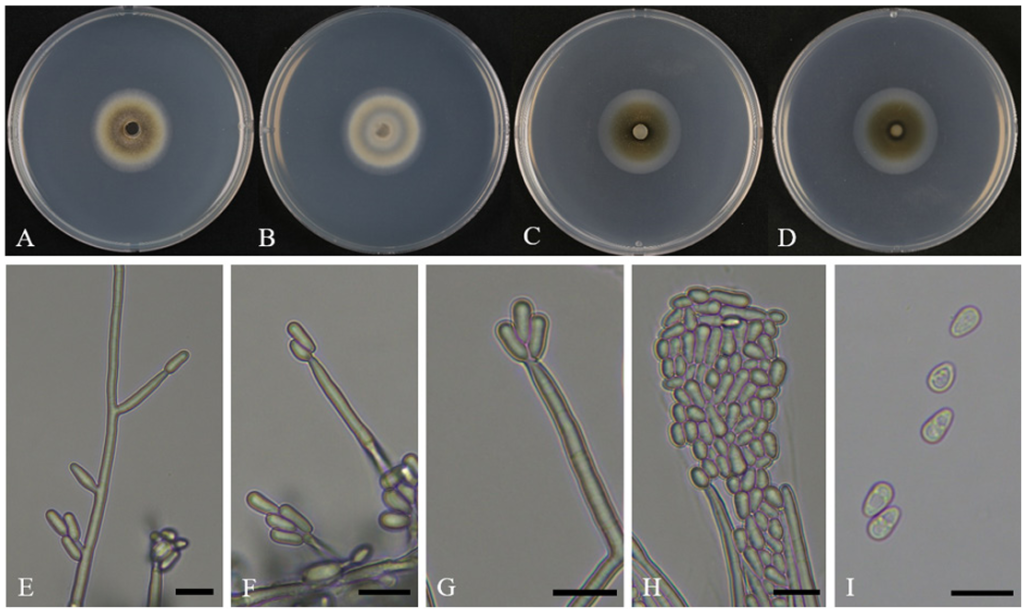
Fig. 1. Cultural and morphological characteristics of ARI-25-A11 (Raffaelea promiscua). A, B: Front and reverse sides of the colony grown on potato dextrose agar (PDA) for 10 days at 25°C. C, D: Front and reverse sides of the colony grown on 2% malt extract agar (MEA) for 10 days at 25°C. E: Branched conidiophores. F–H: Conidiogenous cells producing conidia. I: Conidia. Scale bars: E–I, 10 μm.
Three molecular markers (LSU, SSU, and β-tub) were employed to investigate the molecular and phylogenetic relationships of ARI-25-A11. The lengths of the obtained sequences were 840 bp, 1,022 bp, and 564 bp. These sequences were compared with those of other fungal strains in the NCBI database using a Basic Local Alignment Search Tool (BLAST) search. For the LSU gene, ARI-25-A11 showed the highest similarity, 100.0%, with Raffaelea promiscua PL1001 and R. cyclorhipidii LWP286, followed by 97.2% similarity with R. canadensis CMW 25536 and 94.8% similarity with R. sulalba. For the SSU gene, ARI-25-A11 showed 100.0% similarity with R. promiscua PL1001 and R. canadensis C3169, whereas the similarities with R. cyclorhipidii C2711, R. quercina CBS 147555, and R. ambrosiae CBS 185.64 were 99.4%, 99.1%, and 94.8%, respectively. For the β-tub gene, ARI-25-A11 exhibited 100.0% similarity with both R. promiscua PL1001 and R. promiscua CMW55899ᵀ, while showing a lower similarity of 90.2% with R. canadensis CBS 168.66.
Based on a comparison with sequences registered in the NCBI database, ARI-25-A11 showed high similarity to R. promiscua, R. cyclorhipidii, and R. canadensis, when assessing the LSU and SSU genes, and was found to be most closely related to R. promiscua when assessing the β-tub gene. Notably, all three gene sequences of ARI-25-A11 were nearly identical to those of R. promiscua PL1001, indicating a close genetic relationship. In addition, to determine the phylogenetic position, concatenated sequences of the LSU, SSU, and β-tub genes were used to construct a dataset of 2,426 bp in length. Claviceps fusiformis ATCC 26019 was used as an outgroup for the phylogenetic tree construction. The resulting phylogenetic tree revealed that ARI-25-A11 forms a distinct clade with R. promiscua (Fig. 2).
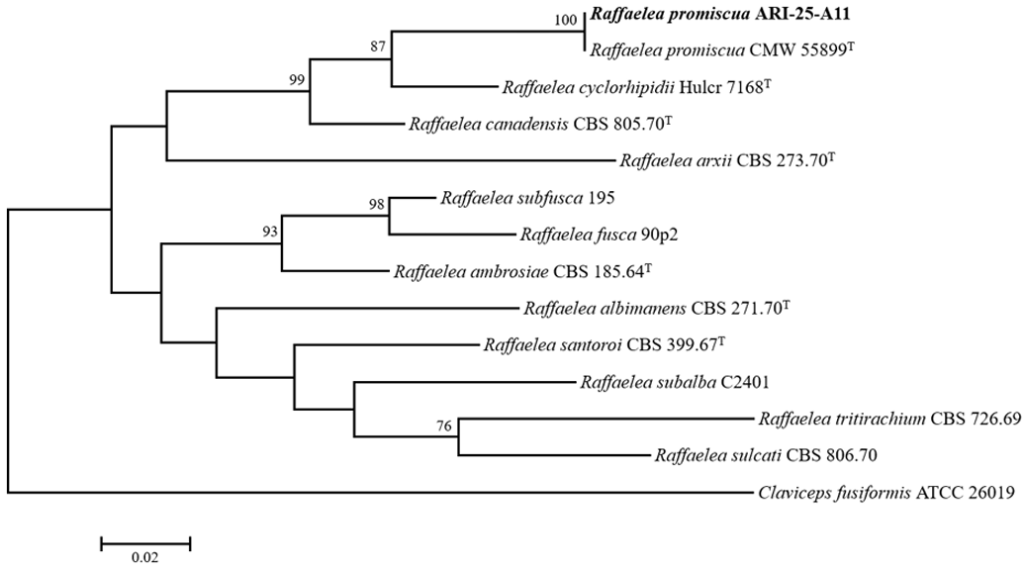
Fig. 2. Maximum-likelihood phylogenetic tree illustrating the placement of strain ARI-25-A11 within the genus Raffaelea, based on concatenated sequences of large subunit ribosomal RNA (LSU), small subunit ribosomal RNA (SSU), and β-tubulin (β-tub) genes. Bootstrap values greater than 70% (from 1,000 replicates) are shown at the corresponding branches. The isolate obtained in this study is highlighted in bold. Claviceps fusiformis (ATCC 26019) was used as the outgroup. The scale bar represents 0.02 nucleotide substitutions per site. T denotes the type strain.
Based on the molecular marker sequences and phylogenetic analyses, ARI-25-A11 was identified as a close relative of Raffaelea promiscua and R. cyclorhipidii. The conidia of ARI-25-A11 were found to be predominantly oblong with swollen upper parts, measuring an average of 5.9 × 3.5 µm, making it morphologically similar to R. promiscua. In contrast, R. cyclorhipidii produced larger, more elongated ellipsoidal conidia with an average size of 7.3 × 3.5 µm, clearly distinguishing it from ARI-25-A11. In culture, ARI-25-A11 formed smooth colonies with aerial hyphae, whereas R. cyclorhipidii produced colonies with tough and wrinkled surfaces, highlighting a notable difference in colony morphology (Table 2).
Table 2. Comparison of the morphological characteristics of strain ARI-24-A11 with those of the reference species Raffaelea promiscua and R. cyclorhipidii
테이블
MEA: malt extract agar; N/A: Not available.
aFungal strain studied in this paper; bSource of description [2]; cSource of description [27].
A comprehensive taxonomic revision of fungal symbionts associated with ambrosia beetles has not been undertaken since Batra [19]. At that time, most known symbionts were classified into anamorphic genera Ambrosiella or Raffaelea based on the belief that these fungi reproduced asexually [20]. Their simplified morphological features made an accurate classification difficult prior to the advent of DNA sequencing technology [21]. Originally, Ambrosiella and Raffaelea were classified based on the proliferation patterns of their conidiogenous cells (annellidic proliferation vs sympodial proliferation). However, subsequent studies revealed no substantial differences in proliferation modes between the two genera, and this morphological criterion was deemed taxonomically unreliable [10].
The taxonomy of Raffaelea has been systematically revised using molecular phylogenetic techniques. Dreaden et al. [6] conducted the first comprehensive multigene phylogenetic analysis of the genus based on LSU, SSU, and β-tub gene sequences. They classified Raffaelea brunnea, R. lauricola, R. scolytodis, R. arxii, R. gnathotrichi, R. fusca, R. subfusca, R. ellipticospora, R. ambrosiae, R. canadensis, R. albimanens, R. subalba, R. tritirachium, R. santoroi, and R. sulcati as Raffaelea sensu stricto, whereas other species were considered to have uncertain taxonomic positions.
A previously unidentified strain, PL1004, was initially identified as R. canadensis by Eskalen and McDonald [22]. However, Dreaden et al. [6] confirmed that this strain is non-pathogenic and genetically distinct from R. canadensis and thus considered it a novel species. In 2021, fungi isolated from an ambrosia beetle were confirmed to be identical to PL1004 and officially described as R. promiscua [2].
In this study, the strain ARI-25-A11 was subjected to phylogenetic analysis based on LSU, SSU, and β-tub gene sequences. Sequence analysis revealed that ARI-25-A11 was identical to PL1004 across all examined loci, and a phylogenetic analysis placed it in the same clade as PL1004. Therefore, strain ARI-25-A11 was considered conspecific with PL1004.
Nel et al. [2] reported that strain CMW 55899, genetically identical to PL1004, is a new species and named it R. promiscua, based on internal transcribed spacer (ITS), LSU, and β-tub gene analyses. In our study, we attempted to sequence the ITS region but were unable to obtain high-quality sequences. This result aligns with previous findings that ITS amplification and interpretation are challenging in Raffaelea [23,24] and that many Raffaelea species lack ITS sequences in the NCBI database.
Accordingly, SSU was used instead of ITS for species identification. However, because Nel et al. [2] did not include the SSU gene when describing R. promiscua as a new species, none of the available R. promiscua strains in the NCBI database had all three genes (LSU, SSU, and β-tub). Therefore, in our analysis, the SSU gene sequence of R. promiscua CMW 55899ᵀ was treated as a gap (missing data), and maximum likelihood analysis was carried out.
In 2022, a taxonomic revision of the order Ophiostomatales, including the genus Raffaelea, was conducted based on four gene regions ITS, LSU, TEF1-α, and RPB2 [25]. However, this revision was considered overly inclusive, and many Raffaelea species lack sequence data for TEF1-α and RPB2, making accurate comparisons difficult. Therefore, in this study, we determined that using LSU, SSU, and β-tub gene sequences remains the most effective approach for identifying species within the genus Raffaelea. Our phylogenetic analysis further supports the validity of this method by enabling effective classification based on these markers. However, a consistent set of molecular markers for species identification has not been established, posing challenges when attempting to compare multiple strains. In particular, sequencing the ITS region remains problematic for many Raffaelea isolates. For example, for R. cyclorhipidii and R. subfusca, both previously reported in Korea, publicly available ITS sequences are lacking [26]. If more isolates of such species could be obtained in the future, it would be intriguing to investigate the intraspecific and intragenomic ITS variations among Raffaelea species distributed in Korea.
The authors declare that they have no potential conflicts of interest.
This study was supported by the “Cooperative Research Program for Agriculture Science and Technology Development (Project No. PJ017183)” funded by the Rural Development Administration, Republic of Korea.
1. Six DL. Ecological and evolutionary determinants of bark beetle-fungus symbioses. Insects 2012;3:339-66. https://doi.org/10.3390/insects3010339 [DOI]
2. Nel WJ, Wingfield MJ, de Beer ZW, Duong TA. Ophiostomatalean fungi associated with wood boring beetles in South Africa including two new species. Antonie van Leeuwenhoek 2021;114:667-86. https://doi.org/10.1007/s10482-021-01548-0 [DOI]
3. Kolařík M, Kirkendall LR. Evidence for a new lineage of primary ambrosia fungi in Geosmithia Pitt (Ascomycota: Hypocreales). Fungal Biol 2010;114:676-89. https://doi. org/10.1016/j.funbio.2010.06.005 [DOI]
4. Bateman C, Huang YT, Simmons DR, Kasson MT, Stanley EL, Hulcr J. Ambrosia beetle Premnobius cavipennis (Scolytinae: Ipini) carries highly divergent ascomycotan ambrosia fungus, Afroraffaelea ambrosiae gen. nov. et sp. nov. (Ophiostomatales). Fungal Ecol 2017;25:41-9. https://doi.org/10.1016/j.funeco.2016.10.008 [DOI]
5. van der Linde JA, Six DL, De Beer WZ, Wingfield MJ, Roux J. Novel ophiostomatalean fungi from galleries of Cyrtogenius africus (Scolytinae) infesting dying Euphorbia ingens. Antonie Van Leeuwenhoek 2016;109:589-601. https://doi.org/10.1007/s10482-016-0661-1 [DOI]
6. Dreaden TJ, Davis JM, de Beer ZW, Ploetz RC, Soltis PS, Wingfield MJ, Smith JA. Phylogeny of ambrosia beetle symbionts in the genus Raffaelea. Fungal Biol 2014;118:970-8. https://doi. org/10.1016/j.funbio.2014.09.001 [DOI]
7. von Arx JA, Hennebert GL. Deux champignons ambrosia. Mycopathol Mycol Appl 1965;25:309-15. https://doi.org/10.1007/BF02049918 [DOI]
8. Kubono T, Ito S. Raffaelea quercivora sp. nov. associated with mass mortality of Japanese oak, and the ambrosia beetle (Platypus quercivorus). Mycoscience 2002;43:255-60. https://doi. org/10.1007/S102670200037 [DOI]
9. Kim KH, Choi YJ, Seo ST, Shin HD. Raffaelea quercus-mongolicae sp. nov. associated with Platypus koryoensis on oak in Korea. Mycotaxon 2009;110:189-97. [DOI]
10. Harrington TC, Fraedrich SW, Aghayeva DN. Raffaelea lauricola, a new ambrosia beetle symbiont and pathogen on the Lauraceae. Mycotaxon 2008;104:399-404. [DOI]
11. Procter M, Nel WJ, Marincowitz S, Crous PW, Wingfield MJ. A new species of Raffaelea from beetle-infested Leucaena leucocephala. Fungal Syst Evol 2020;6:305-14. [DOI]
12. de Beer ZW, Seifert KA, Wingfield MJ. The ophiostomatoid fungi: Their dual position in the Sordariomycetes. In: Seifert KA, de Beer ZW, Wingfield MJ, editors. The ophiostomatoid fungi: Expanding frontiers. CBS biodiversity series, vol. 12. Utrecht, Netherlands: CBSKNAW Fungal Biodiversity Centre; 2013. p. 1-19. [DOI]
13. Vilgalys R, Hester M. Rapid genetic identification and mapping of enzymatically amplified ribosomal DNA from several Cryptococcus species. J Bacteriol 1990;172:4238-46. https://doi. org/10.1128/jb.172.8.4238-4246.1990 [DOI]
14. White TJ, Bruns T, Lee S, Taylor J. Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics. In: Innis MA, Gelfand DH, Sninsky JJ, White TJ, editors. PCR protocols: A guide to methods and applications. New York: Academic Press; 1990. p. 315-22. [DOI]
15. Glass NL, Donaldson GC. Development of primer sets designed for use with the PCR to amplify conserved genes from filamentous ascomycetes. Appl Environ Microbiol 1995;61:1323-30. https://doi.org/10.1128/aem.61.4.1323-1330.1995 [DOI]
16. Kumar S, Stecher G, Tamura K. MEGA7: Molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol Biol Evol 2016;33:1870-4. https://doi.org/10.1093/molbev/ msw054 [DOI]
17. Kimura M. A simple method for estimating evolutionary rates of base substitutions through comparative studies of nucleotide sequences. J Mol Biol 1980;16:111-20. https://doi. org/10.1007/bf01731581 [DOI]
18. Fitch WM. Toward defining the course of evolution: Minimum change for a specific tree topology. Syst Zool 1971;20:406-16. https://doi.org/10.1093/sysbio/20.4.406 [DOI]
19. Batra LR. Ambrosia fungi: A taxonomic revision and nutritional studies of some species. Mycologia 1967;59:976-1017. https://doi.org/10.1080/00275514.1967.12018485 [DOI]
20. Batra LR. Ecology of ambrosia fungi and their dissemination by beetles. Trans Kans Acad Sci 1963;66: 213-36. [DOI]
21. Harrington TC, Aghayeva DN, Fraedrich SW. New combinations in Raffaelea, Ambrosiella, and Hyalorhinocladiella, and four new species from the redbay ambrosia beetle, Xyleborus glabratus. Mycotaxon 2010;111:337-61. [DOI]
22. Eskalen A, McDonald V. First report of Raffaelea canadensis causing laurel wilt disease symptoms on avocado in California. Plant Dis 2011;95:1189. https://doi.org/10.1094/PDIS-0311-0203 [DOI]
23. Harrington TC, Yun HY, Lu SS, Goto H, Aghayeva DN, Fraedrich SW. Isolations from the redbay ambrosia beetle, Xyleborus glabratus, confirm that the laurel wilt pathogen, Raffaelea lauricola, originated in Asia. Mycologia 2011;103:1028-36. https://doi.org/10.3852/10-417 [DOI]
24. Jeyaprakash A, Davison DA, Schubert TS. Molecular detection of the laurel wilt fungus, Raffaelea lauricola. Plant Dis 2014;98:559-64. https://doi.org/10.1094/pdis-08-13-0894-re [DOI]
25. de Beer ZW, Procter M, Wingfield MJ, Marincowitz S, Duong TA. Generic boundaries in the Ophiostomatales reconsidered and revised. Stud Mycol 2022;101:57-120. https://doi. org/10.3114/sim.2022.101.02 [DOI]
26. Jeon MJ, Park S, Jeong JC, Lim J, Han Y, Chi WJ, Kim S. Eight fungal species associated with ambrosia beetles in Korea. Mycobiology 2025;53:1-17. https://doi.org/10.1080/12298093.202 4.2391629
27. Simmons DR, de Beer ZW, Huang YT, Bateman C, Campbell AS, Dreaden TJ, Li Y, Ploetz RC, Black A, Li HF, et al. New Raffaelea species (Ophiostomatales) from the USA and Taiwan associated with ambrosia beetles and plant hosts. IMA Fungus 2016;7:265-73. https:// doi.org/10.5598/imafungus.2016.07.02.06 [DOI]