Lamiya Abasova1,2, In-Young Choi1,2*, Hye-Ryeong Jang1,2, Gyo-Seon Shin3, Ji-Hyun Park3,4*, and Hyeon-Dong Shin5
1Department of Plant Medicine, Jeonbuk National University, Jeonju 54896, Korea
2Research Center for Plant Medicine, Jeonbuk National University, Jeonju 54896, Korea
3Forest Carbon Graduate School, Kookmin University, Seoul 02707, Korea
4Department of Forestry, Environment, and Systems, Kookmin University, Seoul 02707, Korea
5Division of Environmental Science and Ecological Engineering, Korea University, Seoul 02841, Korea
*Correspondence to iychoi@jbnu.ac.kr, jhpark10@kookmin.ac.kr
Korean Journal of Mycology (Kor J Mycol) 2025 September, Volume 53, Issue 3, pages 221-226.
https://doi.org/10.4489/kjm.2025.53.3.9
Received on May 15, 2025, Revised on September 16, 2025, Accepted on September 16, 2025, Published on September 30, 2025.
Copyright © The Korean Society of Mycology.
This is an Open Access article which is freely available under the Creative Commons Attribution-Non-Commercial 4.0 International License (CC BY-NC) (https://creativecommons.org/licenses/by-nc/4.0/).
Erysiphe actinostemmatis is the only known powdery mildew fungus affecting Actinostemma lobatum (Cucurbitaceae) in Japan and China. Anamorph morphology was insufficiently described, and the phylogenetic placement of this species is unknown. To characterize E. actinostemmatis, eight Korean samples collected between 2007 and 2022 were morphologically examined, and the nucleotide sequences of the large subunit (LSU or 28S) gene and internal transcribed spacer (ITS) regions of rDNA were determined using three voucher specimens. Phylogenetic analyses revealed that E. actinostemmatis formed a distinct clade that diverged from the E. aquilegiae clade. In this study, the detailed morphological characterization of the anamorph, including conidial germination patterns and hyphal appressoria, and molecular phylogeny were discussed for the first time, and the sequences of the ITS and LSU genes were provided.
Actinostemma tenerum, conidial germination, Cucurbitaceae, Erysiphe communis, Erysiphe polygoni
Powdery mildew on Actinostemma lobatum Maxim., Franch. & Sav. (syn. Actinostemma tenerum Naudin) had been described as Erysiphe communis (Wallr.) Schltdl. in a broad sense [1]. In 1976, this fungus was treated as E. communis f. cucumidis Marchenko [2], presumably due to its unique parasitism on cucurbitaceous hosts among the wide host range of E. communis. In 1983, Braun [3] raised this fungus to the species level as E. actinostemmatis U. Braun.
According to Braun and Cook [4], the holotype of E. actinostemmatis designated by Braun was collected on October 23, 1910, in Japan and preserved in the National Museum of Nature and Science, Japan (accession no. TNS-F-229705). As the holotype accommodates only the sexual state of the fungus, the original description of E. actinostemmatis was devoid of anamorph characteristics, solely providing the conidial dimension as “approximately 30–38 × 13–17.5 µm”. Furthermore, no sequence data are available for E. actinostemmatis to date. Therefore, the basic information required to characterize E. actinostemmatis as a distinct species in the genus Erysiphe is insufficient for taxonomic purposes.
During our field forays in Korea, eight samples of A. lobatum infected by powdery mildew were collected and preserved at the Korea University Herbarium (KUS). They were labeled as KUS-F22806 (29 Aug 2007, Yangpyeong), F22927 (22 Sep 2007, Yangpyeong), F25657 (7 Oct 2010, Jinju), F27614 (16 Sep 2013, Yangpyeong), F29460 (11 Sep 2016, Imsil), F33083 (20 Jul 2022, Imsil), F33124 (24 Aug 2022, Imsil), F33299 (29 Sep 2022, Imsil), and F33569 (17 Nov 2022, Imsil).
Detailed morphological characteristics of the powdery mildew were examined using an Olympus BX50 microscope (Olympus, Tokyo, Japan). Photomicrographs were captured using a Zeiss AX10 microscope equipped with an AxioCam MRc5 camera (Carl Zeiss, Oberkochen, Germany). All microscopic characterizations and measurements were performed using fresh samples.
Powdery mildew symptoms were observed on the leaves, stems, and fruit (Fig. 1A–F). Chasmothecia were formed in September and October (Fig. 1C–F). The morphological characteristics of powdery mildew are as follows: Hyphae were straight or wavy, hyaline or almost hyaline, septate, branched, and 4–7 µm wide; hyphal appressoria were well-developed, moderately lobed to multi-lobed, and solitary or in opposite pairs (Fig. 1G–I). Conidiophores were 76–120 × 7–9 µm, producing solitary conidia, followed by 2–3 cells; foot-cells of conidiophores were straight to slightly curved, relatively short, and 26–36 µm long (Fig. 1J–K). Conidia were hyaline, oblong-elliptical to ellipsoid, and 30–46 × 15–22 µm, with a length/width ratio of 1.6–2.8 (Fig. 1L). They lacked fibrosin bodies and produced germ tubes in the perihilar position (Fig. 1M–O). Chasmothecia were amphigenous, cauligenous, gregarious to scattered, partly embedded in the mycelial mat, dark brown, spherical, and 90–124 µm in diameter, and contained 4–7 asci per chasmothecium (Fig. 1P–Q). Chasmothecial appendages were mycelioid, about 10–20 in number, septate, not branched, reaching up to four times the chasmothecial diameter, and brown throughout or paler upwards (Fig. 1P). Asci were obovoid, saccate, short-stalked, and 52–64 × 30–38 µm, and contained 6–8 spores per ascus (Fig. 1R). Ascospores were hyaline to subhyaline, oblong-ovoid, and 14–18 × 9–10 µm (Fig. 1S–T). The fungus was initially identified as E. actinostemmatis based on its morphological characteristics, including sexual morph and host specification [4].
To confirm the morphology-based identification and reveal the molecular phylogeny of the fungus, nucleotide sequences of the 5ʹ-end of the large subunit (LSU) gene and the internal transcribed spacer (ITS) region, including 5.8S rDNA gene, of the rDNA were determined from three collections, as outlined by Choi et al. [5]. The forward and reverse sequences were assembled in MEGA11, and consensus sequences were deposited to GenBank under the following accession numbers: PV366477, PV366478, and PV366479 for ITS, and PV366483, PV366484, and PV370038 for LSU. To determine the genetic affinity of our isolates, a comparison was performed against reference sequences using a BLAST search in the NCBI database. Results revealed 100% identity to Pseudoidium hortensiae (MG654731), E. begoniae (MZ958709–MZ95870912), and Erysiphe sp. (EU185636–EU185641) in the ITS, and 99.8% similarity to Pseudoidium pedaliacearum (LC342963, LC342966, LC342967), E. caricae-papayae (LC228614), and
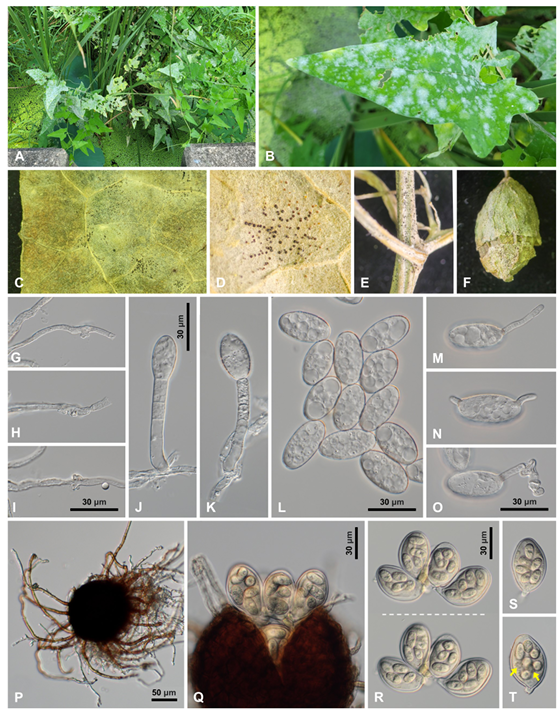
Fig. 1. Erysiphe actinostemmatis, a powdery mildew found on Actinostemma lobatum. A: Powdery mildew symptoms on A. lobatum. B: Close-up view of an infected leaf. C: Chasmothecia formed on the infected leaf. D: Close-up view of the chasmothecia. E: Chasmothecia formed on the stems. F: Chasmothecia formed on the fruit. G–I: Appressoria on the hypha. J–K: Conidiophores. L: Conidia. M–O: Conidia in germination. P: Chasmothecium with appendages. Q: Chasmothecium protruding asci. R: Cluster of asci focused at different depths, showing 6–7 ascospores per ascus. S: Ascus with six ascospores. T: Ascus with eight ascospores. Two ascospores indicated by arrows are out of focus.
E. aquilegiae (LC009942) in the LSU. For phylogenetic analysis, a combined dataset of ITS + LSU was constructed using 34 closely related sequences retrieved from GenBank and aligned using the MUSCLE command in MEGA11 [6]. Erysiphe viburni was used as an outgroup. Phylogenetic trees were generated in PAUP*4.0a using the maximum parsimony method [7] and raxmlGUI 2.0.13 for maximum likelihood (ML) based on the GAMMA model with GTR substitution [8]. The robustness of the internal branches in the resulting trees was tested using bootstrap (BS) analysis through 1,000 replications, and BS values higher than 70% were shown on the relevant branches. Tree scores, including tree length, consistency index, retention index, and rescaled consistency index were also calculated. The final alignment consisted of 38 sequences and 1,349 characters, of which 44 (3.3%) were variable and parsimony-uninformative and 58 (4.3%) were informative for parsimony analysis. The phylogenetic trees generated from the MP and ML analyses were consistent with each other. In the given parsimonious tree, our sequences were clustered with a sequence of Erysiphe sp. on A. lobatum (KY290257) in a distinct branch, which diverged from a clade of the E. aquilegiae complex and was supported by 88/93 BS values in the MP and ML analyses, respectively (Fig. 2). Consequently, following morphological and molecular phylogenetic identification, the
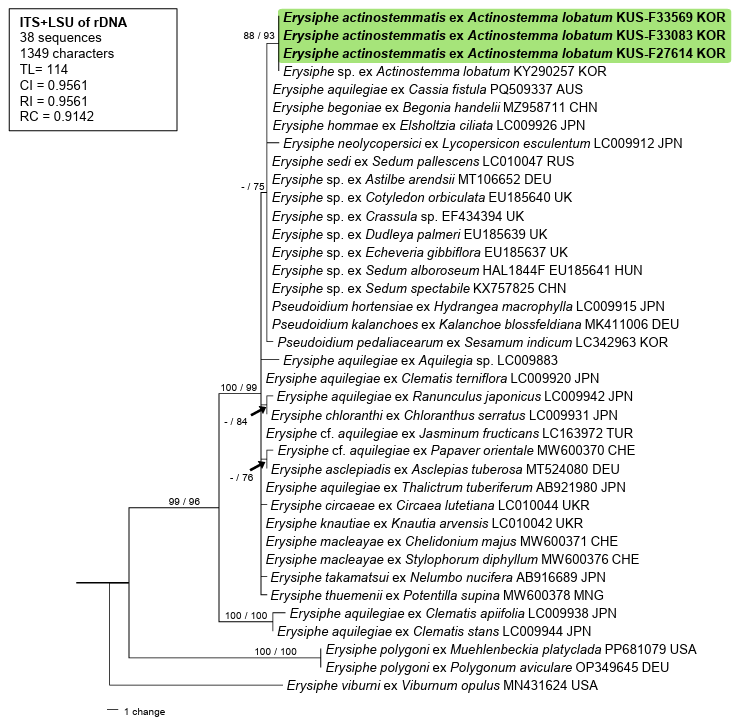
Fig. 2. Parsimonious tree constructed to reveal the phylogeny of Erysiphe actinostemmatis based on a combined dataset of the internal transcribed spacer (ITS) region and the large subunit (LSU) gene of rDNA sequences. The isolates obtained in this study are shown in bold. Bootstrap values (≥70%) obtained from the maximum parsimony and maximum likelihood analyses are indicated on relevant branches, respectively. The calculated tree scores, such as tree length (TL), consistency index (CI), retention index (RI), and rescaled consistency index (RC), are shown in the box.
fungus found on A. lobatum was confirmed to be E. actinostemmatis. In addition, Hong et al. [9] reported the occurrence of an unidentified Erysiphe species on A. lobatum in Korea and provided a sequence for this material (KY290257) that was 674 bp long and represented only the complete sequence of ITS1–5.8S–ITS2 and a partial LSU gene of rRNA. Based on the results of the phylogenetic analyses, we concluded that the sequence of Erysiphe sp. was that of E. actinostemmatis.
To date, E. actinostemmatis has been documented in Japan and China [4,10]. A. lobatum, Schizopepon bryoniifolius Maxim., and Coccinia grandis (L.) Voigt, members of the family Cucurbitaceae, are listed in the host range of E. actinostemmatis [10]. However, the status of E. actinostemmatis ex C. grandis is invalid because of the lack of specimens for examination. Tai reported the presence of E. polygoni DC. on A. lobatum in China [11]. Plants from the family Polygonaceae are the original hosts of E. polygoni globally; however, host jumps of plant pathogens occur frequently. Therefore, to determine the molecular phylogenetic affinity between E. actinostemmatis and E. polygoni, two sequences of the latter species from its original hosts were included in the analyses. These results clearly demonstrated that these two powdery mildew species are not related to each other, neither genetically nor in host specificity.
Sequences of the ITS (KP400511, KP400512) region and the LSU (KP337407, KP202686) gene obtained from powdery mildew isolates (DUCC510 and DUCC511) on A. lobatum were found to be submitted to GenBank as E. aquilegiae. These sequences were manually compared with our sequences, and four nucleotide differences were found in the ITS regions; however, the sequences of the LSU gene were identical to ours. In our initial analyses conducted to determine the relationship with E. aquilegiae, these sequences were grouped with E. aquilegiae. Considering the probability of host misidentification, these sequences were excluded from the final phylogenetic analyses.
The genus Actinostemma Griff. (Cucurbitaceae) consists of herbaceous plants native to East Asia (https://powo.science.kew.org). A. lobatum has recently been identified as a promising source of antifungal substances. Its butanol extracts are highly effective in suppressing the growth of various fungal plant pathogens including Magnaporthe oryzae, which causes rice blast disease [12]. Several species, including E. actinostemmatis, Golovinomyces cucurbitacearum, G. orontii, and Podosphaera xanthii, are major powdery mildew pathogens on cucurbitaceous plants, of which only E. actinostemmatis has been confirmed to infect Actinostemma species. To our knowledge, this is the first report of E. actinostemmatis from Korea. Detailed morphological characterization of its anamorph, including germination patterns, and molecular phylogeny were discussed, and sequences of the ITS and LSU of rDNA were provided.
The authors declare no conflicts of interest.
This research was supported by the Global-Learning & Academic research institution for Master’s·PhD students, and Postdocs (LAMP) Program of the National Research Foundation of Korea (NRF) grant funded by the Ministry of Education (No. RS-2024-00443714) and by the R&D Program for Resolving Current Issues in Forest Disaster and Damage (No. RS-2024-00403034) funded by the Korea Forest Service (Korea Forestry Promotion Institute).
1. Amano K. Host range and geographical distribution of the powdery mildew fungi. Tokyo, Japan: Japan Scientific Societies Press; 1986.
2. Marchenko PD. Novi formi Erysiphaceae znaydeni v zakhidnikh oblastjakh URSR. Ukr Bot Zhur 1976;33:271-6.
3. Braun U. Descriptions of new species and combinations in Microsphaera and Erysiphe (IV). Mycotaxon 1983;18:113-29. [DOI]
4. Braun U, Cook RTA. Taxonomic manual of the Erysiphales (powdery mildews). CBS biodiversity series 11. Utrecht, Netherlands: Centraalbureau voor Schimmelcultures; 2012.
5. Choi IY, Abasova L, Choi JH, Park JH, Shin HD. Erysiphe cornicola, a powdery mildew occurring on Cornus controversa in Korea. Korean J Mycol 2023;51:57-62. [DOI]
6. Tamura K, Stecher G, Kumar S. MEGA11: Molecular evolutionary genetics analysis version 11. Mol Biol Evol 2021;38:3022-7. https://doi.org/10.1093/molbev/msab120 [DOI]
7. Swofford DL. PAUP: Phylogenetic analyses using parsimony (and other methods) 4.0b10. Sunderland: Sinauer Associates; 2002.
8. Silvestro D, Michalak I. raxmlGUI: A graphical front-end for RAxML. Org Divers Evol 2012;12:335-7. https://doi.org/10.1007/s13127-011-0056-0 [DOI]
9. Hong SH, Choi IY, Kwon JH, Cho SE, Shin HD. First report of powdery mildew caused by an Erysiphe sp. on Actinostemma lobatum in Korea. Plant Dis 2017;101:841. https://doi. org/10.1094/PDIS-12-16-1734-PDN [DOI]
10. Farr DF, Rossman AY. Fungal databases [Internet]. Beltsville, MD: United States Department of Agriculture, Agricultural Research Service; 2025 [cited 2025 Mar 16]. Available from: https://nt.ars-grin.gov/fungaldatabases/.
11. Tai FL. Sylloge fungorum sinicorum. Beijing, China: Science Press; Academia Sinica; 1979.
12. Choi S, Lee SH, Hwang BS, Oh YT, Jeon J. Antifungal activity-guided analysis of Actinostemma lobatum extracts through serial sub-fractions. Plant Pathol J 2024;40:218-24. https://doi.org/10.5423/ppj.nt.11.2023.0152 [DOI]