Yu-Na Choi, Ji-Won Kim, Eun-Ju Kim, and Ahn-Heum Eom*
Department of Biology Education, Korea National University of Education, Cheongju 28173, Korea
*Correspondence to eomah@knue.ac.kr
Korean Journal of Mycology (Kor J Mycol) 2025 September, Volume 53, Issue 3, pages 227-235.
https://doi.org/10.4489/kjm.2025.53.3.10
Received on August 26, 2025, Revised on September 18, 2025, Accepted on September 18, 2025, Published on September 30, 2025.
Copyright © The Korean Society of Mycology.
This is an Open Access article which is freely available under the Creative Commons Attribution-Non-Commercial 4.0 International License (CC BY-NC) (https://creativecommons.org/licenses/by-nc/4.0/).
Endophytic fungi colonize asymptomatic healthy plant tissues and often produce bioactive compounds. In this study, two endophytic fungi newly recorded in Korea were isolated from the roots of Rhododendron mucronulatum and R. schlippenbachii collected from Mungyeong: Pezicula radicicola, and Codinaea gonytrichoides. Pure cultures were obtained from surface-sterilized root segments and characterized using morphological features and multilocus phylogenetic analyses of the internal transcribed spacer (ITS) region, large subunit ribosomal RNA (LSU) region, and RNA polymerase II second largest subunit (RPB2) regions. Strain KNUE25E045 was identified as P. radicicola and strain KNUE25E068 as C. gonytrichoides. This study provides the first record of P. radicicola and C. gonytrichoides in Korea, thereby expanding the known diversity of root-associated endophytic fungi in ericaceous plants and highlighting the need for further studies on their ecological roles.
Codinaea gonytrichoides, Endophytic fungi, Pezicula radicicola, Rhododendron
The genus Rhododendron (Ericaceae), comprising more than 800 species worldwide, is one of the most valuable ornamental groups cultivated for horticultural purposes for over two centuries. It is an ecologically important component of temperate ecosystems, often dominating acidic and nutrient-poor soils [1,2]. In Korea, the family Ericaceae consists of eight genera and 32 species, including varieties, cultivars, and hybrids, with Rhododendron representing the largest genus, encompassing 20 species [3]. In addition to horticultural purposes, several species of Rhododendron have been reported to be medicinal plants [4]. Thus, Rhododendron is not only an economically important plant, but also an ecologically significant component of temperate forests and alpine ecosystems.
Endophytic fungi are microorganisms that inhabit living plant tissues without causing apparent disease symptoms [5]. They are widely distributed in terrestrial ecosystems and are closely associated with diverse plant hosts. Their ecological roles are diverse, and they function as mutualists, saprobes, or latent pathogens depending on the host and environmental conditions [6,7]. Many endophytes produce secondary metabolites with biological activities, thereby contributing to host growth, stress tolerance, and pathogen resistance [8,9].
Endophytes have drawn considerable attention, particularly in ericaceous plants, because of their high frequency and bioactivity. More than 30% of the endophytes isolated from Ericaceae have been reported to exhibit antimicrobial or antifungal activities, underscoring their importance in both ecological interactions and applied research [9]. However, despite the ecological and economic significance of Rhododendron, studies on the diversity of its root-associated endophytes in Korea are scarce [10]. Clarifying their taxonomic diversity is essential for understanding their ecological functions and assessing their potential as novel sources of bioactive compounds.
During a survey of the root-associated endophytic fungi of ericaceous plants in Korea, two previously unrecorded species, Pezicula radicicola and Codinaea gonytrichoides, were identified and are reported in this study.
In April 2025, the root samples of Rhododendron mucronulatum Turcz. and Rhododendron schlippenbachii Maxim. were collected from Mungyeong, Korea (36° 39′ 44.49″ N, 128° 10′ 33.29″ E and 36° 39′ 41.95″ N, 128° 10′ 35.63″ E, respectively). Only asymptomatic and undamaged samples were selected for further analysis. The samples were placed in sterile polyethylene bags, transported to the laboratory within 12 hours, and immediately subjected to surface sterilization.
The root samples were thoroughly rinsed under running tap water and cut into approximately 2-mm segments. The segments were surface sterilized by immersion in 95% ethanol for 30 s, followed by rinsing with sterile distilled water [10–12]. Sterilized segments were placed on potato dextrose agar (PDA; Difco Laboratories, Detroit, MI, USA) and incubated at 25°C in the dark. Emerging hyphae were subcultured on fresh PDA to obtain pure isolates.
Pure isolates were cultured on malt extract agar (MEA; Kisan Bio, Seoul, Korea) and PDA at 25°C in darkness. Colony morphology, including pigmentation, texture, and margin characteristics, was recorded after seven days. Microscopic features of the reproductive structures were examined using a light microscope (Axio Imager A2, Carl Zeiss, Oberkochen, Germany).
Genomic DNA was extracted from fresh mycelia using a HiGene Genomic DNA Prep Kit (Biofact, Daejeon, Korea). The internal transcribed spacer (ITS) region, large subunit ribosomal RNA (LSU) region, and RNA polymerase II second largest subunit (RPB2) gene were amplified using primer pairs ITS1F/ ITS4 [13], LR0R/LR5 [14], and fRPB2-5F/fRPB2-7cR [15], respectively. PCR products were sequenced by Solgent Co., Ltd. (Daejeon, Korea). The resulting sequences were compared to those available in the National Center for Biotechnology Information (NCBI) database using the Basic Local Alignment Search Tool (BLAST).
The analysis incorporated the combined sequences of ITS, LSU, and RPB2, as applicable. Phylogenetic trees were constructed using maximum likelihood in the MEGA11 software [16]. The evolutionary models chosen for the analysis were TN93+I and TN93+G+I for strains KNUE25E045 and KNUE25E068, respectively. Bootstrapping was performed with 1000 replicates to assess the robustness of the tree topology. The isolated strains were deposited at the National Institute of Biological Resources (NIBR), and their sequences were registered with NCBI under the accession numbers.
Phylogenetic analyses of the ITS, LSU, and RPB2 sequences confirmed the identification of the isolated fungal strains. For strain KNUE25E045, the ITS sequence exhibited 99.64% identity with Pezicula radicicola isolate 102 (GenBank accession OP895794), the LSU sequence showed 99.77% identity with P. radicicola VKM F-4856 (GenBank accession MT774100), and the RPB2 sequence was 99.74% identity with P. radicicola NRRL 12192 (GenBank accession MH805867). In the maximum likelihood tree constructed from the combined ITS and LSU sequences, KNUE25E045 clustered with P. radicicola CBS 112868 (Fig. 1), confirming its identification. Additionally, a maximum likelihood tree constructed from the RPB2 sequences showed that KNUE25E045 clustered with P. radicicola NRRL 12192, further supporting its identification (Fig. 2). Strain KNUE25E068 showed 100% ITS identity to Codinaea gonytrichoides CBS 593.93 (GenBank accession AF178556), and its LSU sequence was 99.88% identical to C. gonytrichoides NN076206 (GenBank accession OL655160). Phylogenetic analysis of the combined ITS and LSU sequences placed KNUE25E068 in a distinct clade with C. gonytrichoides CFCC 54456 (Fig. 3), supporting its classification.
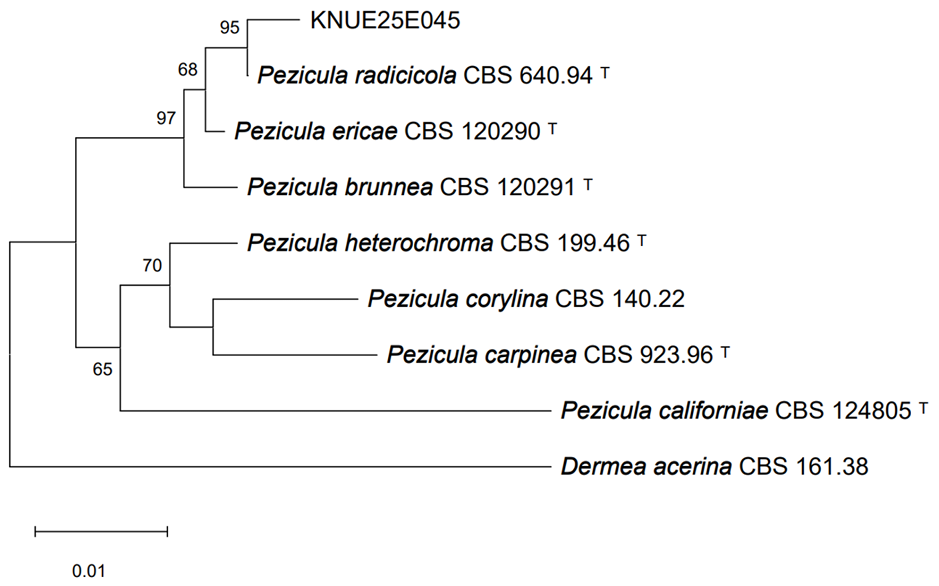
Fig. 1. Maximum likelihood tree of Pezicula radicicola KNUE25E045 based on concatenated internal transcribed spacer (ITS) and large subunit ribosomal RNA (LSU) sequences. Dermea acerina was the outgroup. Bootstrap values > 50% (1,000 replicates) are shown at nodes.ᵀ indicates ex-type cultures.
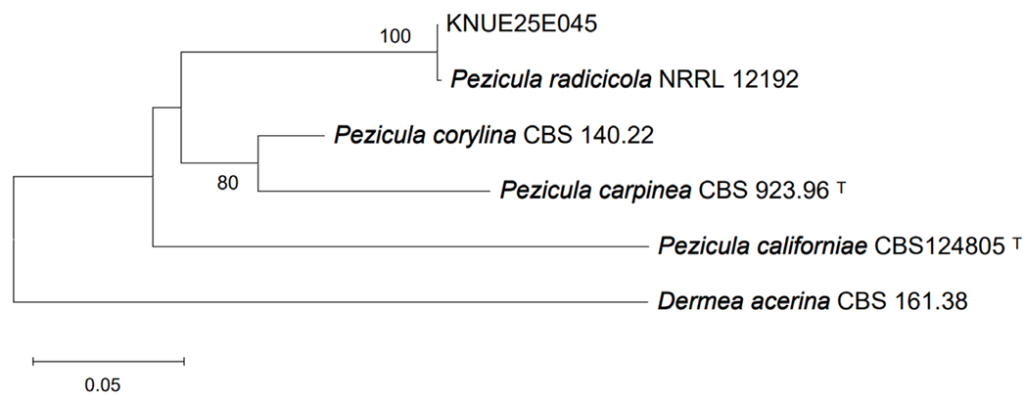
Fig. 2. Maximum likelihood tree of Pezicula radicicola KNUE25E045 based on RNA polymerase II second largest subunit (RPB2) sequences. Dermea acerina was the outgroup. Bootstrap values > 50% (1,000 replicates) are shown at nodes. ᵀ indicates ex-type cultures.
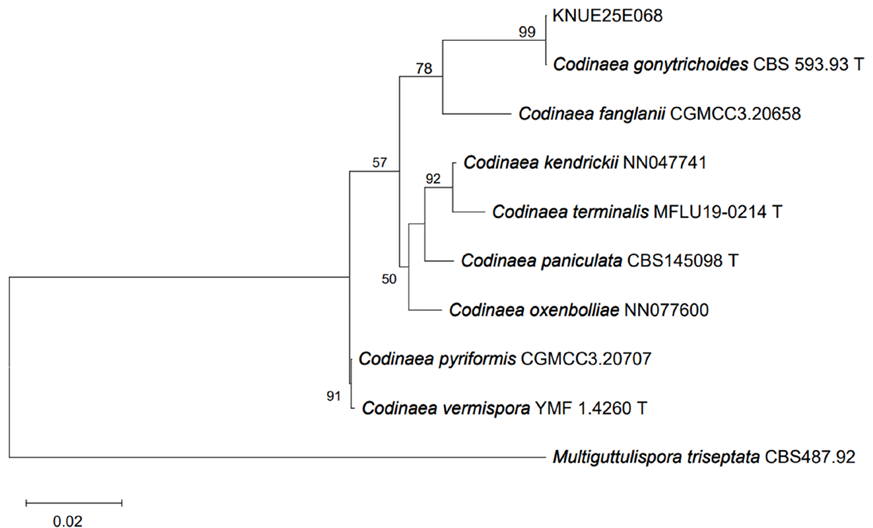
Fig. 3.Maximum likelihood tree of Codinaea gonytrichoides KNUE25E068 based on concatenated internal transcribed spacer (ITS) and large subunit ribosomal RNA (LSU) sequences. Multiguttulispora triseptata was the outgroup. Bootstrap values > 50% (1,000 replicates) are shown at nodes. ᵀ indicates ex-type cultures.
Pezicula radicicola (T. Kowalski & C. Bartnik) P.R. Johnst., IMA Fungus 5 (1): 104 (2014) [MB#808969] (Fig. 4).
Morphological characteristics of strain KNUE25E045: After six days of incubation at 25°C, colonies on PDA reached a diameter of 20.0–25.0 mm. The colony surface was generally brown, with a distinct white margin forming an almost circular outline, even in appearance, and exhibited a cottony texture. The reverse side was predominantly brown with a white margin. On MEA, the colonies reached a diameter of 16.0–21.0 mm. The colony surface was yellowish-white and nearly circular, with a surface and texture similar to those observed on PDA. The reverse was dark reddish-brown at the center and gradually became yellowish-white toward the margins. The conidiophores are solitary, straight to slightly curved, and hyaline. Conidia are aseptate, typically curved, measuring (18.2–)22.7(–26.5) × (5.2–)7.4(–8.6) μm (n = 20).
Specimen examined: Mungyeong, Gyeongsangbukdo, Korea, 36° 39′ 44.49″ N, 128° 10′ 33.29″ E, April 25, 2025, P. radicicola, isolated from R. mucronulatum, strain KNUE25E045, NIBRFGC000514081, GenBank No. PX138773 (ITS), PX138774 (LSU), and PX206067 (RPB2).
Notes: This species was originally isolated from the fine roots of Quercus robur L. and was described as Cryptosporiopsis radicicola by Kowalski & Bartnik, 1995 [17]. The anamorphs of many species formerly described under the genus Cryptosporiopsis have been shown to belong to Pezicula, which is now treated as its synonym [18,19]. Species of Pezicula have frequently been reported in temperate regions worldwide, where they occur either as saprobes on recently dead woody substrates or as endophytes in living branches and roots [17,20]. Strain KNUE25E045 was isolated as an endophyte from the roots of R. mucronulatum, which is consistent with the ecological roles previously reported for members of this genus. Most Pezicula species produce both macroconidia and microconidia during the asexual stage on natural substrates, as well as in culture [21]. However, in strain KNUE25E045, only macroconidia were observed under the culture conditions, and further observations are expected to provide more detailed morphological information.
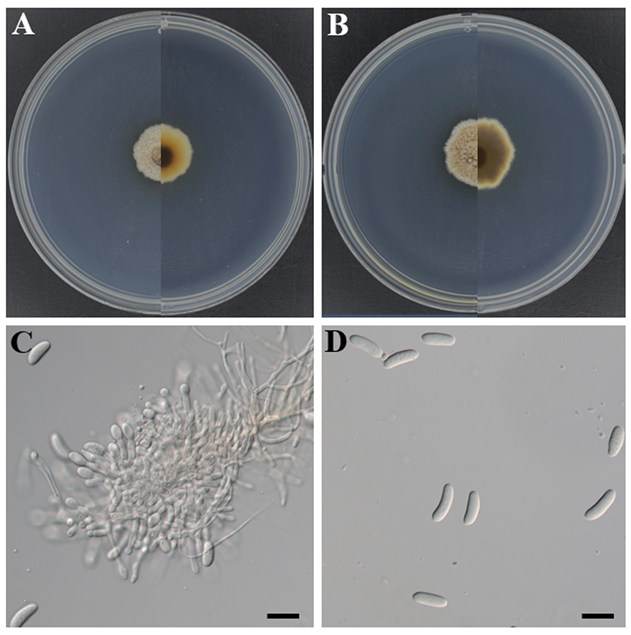
Fig. 4. Morphology of Pezicula radicicola KNUE25E045. A and B: Colonies after seven days of growth at 25°C, showing the obverse (left) and reverse (right) sides on malt extract agar (A) and potato dextrose agar (B). C: Conidiophores. D: Conidia. Scale bars: C and D, 20 μm.
Codinaea gonytrichoides Shearer & J.L. Crane, Mycologia 63 (2): 245 (1971) [MB#372655] (Fig. 5).
Morphological characteristics of strain KNUE25E068: After seven days of incubation at 25°C in the dark, colonies on PDA reached a diameter of 18.0–22.0 mm. The colony surface was gray with a white margin forming at the edges, and the colony shape was irregular with a cottony texture. The reverse side of the colony was dark brown, with a white margin. On MEA, the colonies reached a diameter of 23.0–28.0 mm. The surfaces of the colonies were white, irregular in shape, and cottony in texture. The reverse side was black at the center and gradually faded to white toward the margin. Conidiophores arose laterally from the hyphae, with the basal portion dark brown and gradually becoming subhyaline toward the apex. Phialides occasionally originate from node-like structures that form one or multiple units, each characterized by a funnel-shaped (collarette infundibuliform) morphology. The conidia are falcate and tapered at both ends. They were aseptate and hyaline with straight or slightly curved setulae at both ends. The size of the spores was (12.1−)13.9(−15.8) × (2.6−)3.0(−3.5) μm (n = 20).
Specimen examined: Mungyeong, Gyeongsangbukdo, Korea, 36° 39′ 41.95″ N, 128° 10′ 35.63″ E, April 25, 2025, C. gonytrichoides, isolated from R. schlippenbachii, strain KNUE25E068, NIBRFGC000514082, GenBank No. PX138822 (ITS), and PX138823 (LSU).
Notes:
This species has been reported as a saprobe on decaying leaves, seeds, herbaceous stems, and other unidentified hosts and occurs in diverse habitats, including both freshwater and terrestrial environments. In agreement with previous descriptions, C. gonytrichoides was distinguished from other species of Codinaea by the presence of nodose hyphae at the septa, giving rise to lateral phialides [22,23], a feature clearly observed in strain KNUE25E068 (Fig. 5C). The falcate and aseptate conidia bear characteristic setulae at both ends, which is a diagnostic feature important for classification within the genus Codinaea (Fig. 5D) [24]. Strain KNUE25E068 is the first record of the genus Codinaea in Korea.
In this study, two endophytic fungi, Pezicula radicicola and Codinaea gonytrichoides, were isolated from the roots of Rhododendron spp. in Korea and identified based on their morphological characteristics and multilocus phylogenetic analyses. This is the first report of these species in Korea, expanding the known diversity of root-associated fungi in ericaceous hosts. The present study provides a taxonomic basis for future studies. Further research is needed to elucidate the ecological roles and functional significance of these fungi in their host plants.
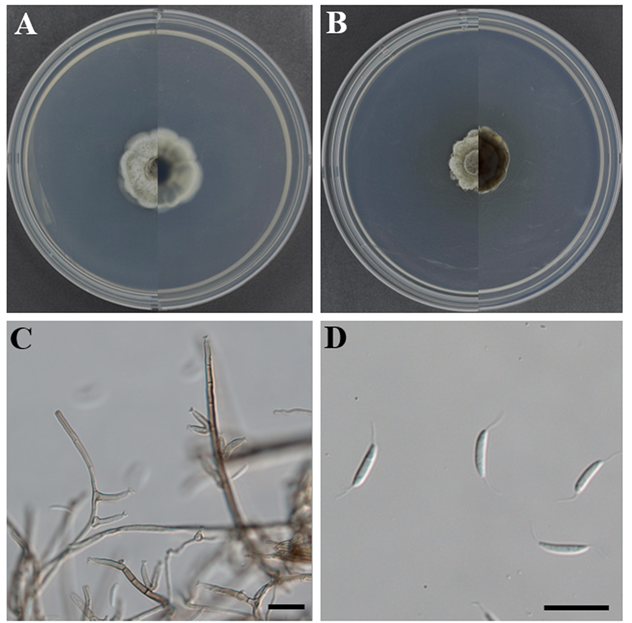
Fig. 5. Morphology of Codinaea gonytrichoides KNUE25E068. A and B: Colonies after seven days of growth at 25°C, showing the obverse (left) and reverse (right) sides on malt extract agar (A) and potato dextrose agar (B). C: Conidiophores. D: Conidia. Scale bars: C and D, 20 μm.
The authors declare no conflicts of interest.
This work was supported by a grant from the National Institute of Biological Resources (NIBR202502103), funded by the ministry of Environment (MOE) of the Republic of Korea .
1. Krebs SL. Rhododendron. In: Van Huylenbroeck J, editor. Ornamental crops. 2nd ed. Cham, Switzerland: Springer; 2018. p. 673-718.2 [DOI]
2. Basnett S, Ganesan R. A comprehensive review on the taxonomy, ecology, reproductive biology, economic importance and conservation status of Indian Himalayan Rhododendrons. Bot Rev 2022;88:505-44. https://doi.org/10.1007/s12229-021-09273-z3 [DOI]
3. National Institute of Biological Resources (NIBR). National species information system [Internet]. Incheon: NIBR; 2025 [cited 2025 August 18]. Available from: https://species.nibr.go.kr.4
4. Min JH, Kim JH, Lee SI, Jeong JG. A herbological study on the plants of Eriaceae in Korea. Kor J Herbology 2013;28:41-8. https://doi.org/10.6116/kjh.2013.28.4.41.5 [DOI]
5. Stone JK, Polishook JD, White JF. Endophytic fungi. In: Mueller GM, Bills GF, Foster MS, editors. Biodiversity of fungi: Inventory and monitoring methods. Burlington, MA: Elsevier; 2004. p. 241-70.6 [DOI]
6. Hirsch G, Braun U. Communities of parasitic microfungi. In: Winterhoff W, editor. Handbook of vegetation science. Volume 19. Fungi in vegetation science. Dordrecht, Netherlands: Kluwer Academic Publishers; 1992. p. 225-50.6 [DOI]
7. Saikkonen K, Faeth SH, Helander M, Sullivan TJ. Fungal endophytes: A continuum of interactions with host plants. Annu Rev Ecol Evol Syst 1998;29:319-43. https://doi.org/10.1146/annurev.ecolsys.29.1.3198 [DOI]
8. Schulz B, Sucker J, Aust HJ, Krohn K, Ludewig K, Jones PG, Döring D. Biologically active secondary metabolites of endophytic Pezicula species. Mycol Res 1995;99:1007-15. https://doi.org/10.1016/S0953-7562(09)80766-19 [DOI]
9. Fisher PJ, Anson AE, Petrini O. Antibiotic activity of some endophytic fungi from ericaceous plants. Bot Helv 1984;94:249-53.10.
10. Park H, Eom AH. Characterization of Rhizodermea veluwensis isolated from the roots of Rhododendron mucronulatum in Korea. Kor J Mycol 2017;45:102-6. https://doi.org/10.4489/KJM.2017001211. [DOI]
11. Sieber TN. Fungal root endophytes. In: Waisel Y, Eshel A, Beeckman T, Kafkafi U, editors. Plant roots: The hidden half. 3rd ed. Boca Raton, FL: CRC Press; 2002. p. 887-917.12 [DOI]
12. Hambleton S, Currah RS. Fungal endophytes from the roots of alpine and boreal Ericaceae. Can J Bot 1997;75:1570-81. https://doi.org/10.1139/b97-86913 [DOI]
13. Gardes M, Bruns TD. ITS primers with enhanced specificity for basidiomycetes-application to the identification of mycorrhizae and rusts. Mol Ecol 1993;2:113-8. https://doi.org/10.1111/j.1365-294x.1993.tb00005.x14 [DOI]
14. White TJ, Bruns TD, Lee SB, Taylor JW. Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics. In: Innis MA, Gelfand DH, Sninsky JJ, editors. PCR protocols: A guide to methods and applications. New York, NY: Academic Press; 1990. p. 315-22.15 [DOI]
15.1Liu YJ, Whelen S, Hall BD. Phylogenetic relationships among ascomycetes: Evidence from an RNA polymerase II subunit. Mol Biol Evol 1999;16:1799-808. https://doi.org/10.1093/oxfordjournals.molbev.a02609216 [DOI]
16. Tamura K, Stecher G, Kumar S. MEGA11: Molecular evolutionary genetics analysis version 11. Mol Biol Evol 2021;38:3022-7. https://doi.org/10.1093/molbev/msab120 [DOI]
17. Kowalski T, Bartnik C. Cryptosporiopsis radicicola sp. nov. from roots of Quercus robur. Mycol Res 1995;99:663-6. https://doi.org/10.1016/S0953-7562(09)80524-818 [DOI]
18. Johnston PR, Seifert KA, Stone JK, Rossman AY, Marvanová L. Recommendations on generic names competing for use in Leotiomycetes (Ascomycota). IMA Fungus 2014;5:91-120. https://doi.org/10.5598/imafungus.2014.05.01.1119 [DOI]
19. Chen C, Verkley GJM, Sun G, Groenewald JZ, Crous PW. Redefining common endophytes and plant pathogens in Neofabraea, Pezicula, and related genera. Fungal Biol 2016;120:1291-322. https://doi.org/10.1016/j.funbio.2015.09.01320 [DOI]
20. Kochkina GA, Ivanushkina NE, Pinchuk IP, Ozerskaya SM. Endophytic fungi Pezicula radicicola in the root nodules of actinorhizal plants. Microbiology 2022;91:750-6. https://doi.org/10.1134/S002626172260162221 [DOI]
21. Verkley GJM. A monograph of the genus Pezicula and its anamorphs. Stud Mycol 1999;44:1-176.22
22. Shearer CA, Crane JL. Fungi of the Chesapeake Bay and its tributaries. I. Patuxent River. Mycologia 1971;63:237-60. https://doi.org/10.1080/00275514.1971.1201910623 [DOI]
23. Réblová M, Kolařík M, Nekvindová J, Réblová K, Sklenář F, Miller AN, Hernández-Restrepo M. Phylogenetic reassessment, taxonomy, and biogeography of Codinaea and similar fungi. J Fungi 2021;7:1097. https://doi.org/10.3390/jof712109724 [DOI]
24. Réblová M. The genus Chaetosphaeria and its anamorphs. Stud Mycol 2000;45:149-68. [DOI]