Hyeok Park*, Jeomsoon Kim, and Sumi Kim
Highland Agriculture Research Institute, National Institute of Crop Science, Pyeongchang 25342, Korea
*Correspondence to mycoph@korea.kr
Korean Journal of Mycology (Kor J Mycol) 2024 December, Volume 52, Issue 4, pages 257-263.
https://doi.org/10.4489/kjm.520404
Received on November 01, 2024, Revised on December 02, 2024, Accepted on December 02, 2024, Published on Dec 30, 2024.
Copyright © The Korean Society of Mycology.
This is an Open Access article which is freely available under the Creative Commons Attribution-NonCommercial 4.0 International License (CC BY-NC) (https://creativecommons.org/licenses/by-nc/4.0/).
In the present study, we isolated endophytic fungi from the leaves of spring-cultivated potato plants. The fungal strains were identified using the morphological characteristics and molecular analysis of the internal transcribed spacer, large subunit of rDNA, and β-tubulin gene. We confirmed the presence of two endophytic fungal species not previously recorded in Korea: Didymella sinensis and Sagenomella oligospora. Here, we describe the morphological and phylogenetic characteristics of these two fungal species.
Agriculture, Crop, Fungal endophyte, Potato, Symbiosis
Endophytic fungi live in all plant tissues, including leaves, stems, and roots, without causing any symptoms [1]. They play a role in chemically protecting host plants from external pathogens by secreting secondary metabolites [2]. The host range of endophytic fungi is diverse, ranging from woody plants [3] to bryophytes [4], and they form symbiotic relationships with crops [5].
Potatoes (Solanum tuberosum L.) belong to the family Solanaceae and are one of the most important food crops worldwide. In Korea, potatoes cultivated in spring are the main crop type and account for 63% of the total potato production [6,7]. In the spring cultivation of potatoes, the management of diseases, such as early blight, late blight, and Fusarium wilt, is important; therefore, endophytic fungi that can control potato diseases are useful. Some studies have been conducted on endophytic fungi isolated from potatoes and these studies refer to the role of endophytic fungi in disease control in potatoes [8–10]. In this study, we report two previously unrecorded endophytic fungi isolated from potato leaves during spring potato cultivation in Korea.
Plant samples of spring-cultivated potatoes were collected in June 2024. Healthy potato leaves without disease symptoms were collected from the test fields in Chungcheongbuk-do Agricultural Research and Extension Services (36°43’34.3″N, 127°27’58.6″E) and Gyeongsangnam-do Agricultural Research and Extension Services (35°12’37.8″N, 128°07’02.7″E). The samples were transported to the laboratory within 24 h. Potato leaves were surface-sterilized in 1% NaOCl for 1 min and in 70% EtOH for 2 min, and then plated on water agar (WA) medium [11]. With culturing at 25℃ on the dark side, mycelium or single conidium were subcultured onto potato dextrose agar (PDA) and malt extract agar (MEA). The morphology of the fungal strains was observed using a dissecting microscope, and the morphological characteristics of the conidia were observed using an optical microscope (Table 1). For the molecular identification of the fungal species, genomic DNA was extracted using the DiaStar™ Direct Multiplex Polymerase Chain Reaction (PCR) Kit (Solgent Co., Ltd., Daejeon, Korea). The internal transcribed spacer (ITS) region of rDNA was amplified using the fungi-specific primers ITS1F/ITS4 [12], the large subunit (LSU) region was amplified using the primers LR0R/LR16 [13], and the β-tubulin DNA (TUB) region was amplified using the primers Bt2a/Bt2b [14], respectively. The PCR products were electrophoresed on a 1.5% agarose gel for 20 min, each DNA sequence fragment was confirmed, and Sanger DNA sequencing was performed (Bionics Co., Ltd., Seoul, Korea). DNA sequences were analyzed to search for the most matching species using Basic Local Alignment Search Tool (BLAST) at the National Center for Biotechnology Information (NCBI), and phylogenetic analysis was performed by creating a neighbor-joining (NJ) phylogenetic tree with concatenated DNA sequences using the MEGA 7.0.26 program [15]. The analyzed DNA sequences were registered in the NCBI GenBank database, and the identified fungal strains were deposited in the Korean Agricultural Culture Collection (KACC) of the National Institute of Agricultural Sciences.
Table 1. Morphological characteristics of endophytic fungal strains isolated in this study
| Strains | Colonies | Conidia | |||
|---|---|---|---|---|---|
| Cultural condition | Color | Size | Shape | ||
| Didymella sinensis | |||||
| Chen et al. [16] | MEA, 25℃, 7 d | Greyish brown on both sides | 57–60 mm in diam. | Regular margins | Undescribed |
| Strain HARI24E001 | MEA, 25℃, 7 d | Dark brown, covered with white fluffy aerial mycelium; reverse dark brown in center, light brown in margins | (In three-point cultured plate) 34–39 mm in diam. | Raised colony, regular margins with radial mycelium growth | Hyaline chlamydospore, globose to subglobose, (9.69–) 13.71 (–16.70) × (7.94–) 11.09 (–14.76) μm in diam. |
| PDA, 25℃, 7 d | Light gray; reverse light grey ish-brown in center, light gray in margins | (In three-point cultured plate) 24–27 mm in diam. | Flat colony, highly irregular margins, mycelia digging into the substrate | ||
| Sagenomella oligospora | |||||
| Gams [19] | MEA, 20 –25℃, 15 d | Initially white, pale olivac eous-gray as conidia mature | 24–36 mm in diam. | Thinly floccose, pro strate hyphae | Warts outside the thick spore wall, subglobose, tapering apical and base (including the connectives) 6.5–8.5 ×4.5–6.0 μm |
| Strain HARI24E013 | MEA, 25℃, 7 d | Pale beige; reverse dark beige, yellowish-brown as conidia pigmented in center of colony | (In 3–point cultured plate) 9–11 mm in diam. | Flat colony, irregular margins, slow mycelial growth | Bright yellowish-brown, ovoid or fusiform, warts outside the thick spore wall, cicatrix of conidiophore segmentation in apical and base (not included the connectives) (3.92–) 4.58 (–5.52) × (1.91–) 3.18 (–3.88) μm in diam. |
| PDA, 25℃, 7 d | Bright white; reverse beige, reddish brown in center | (In three-point cultured plate) 14–15 mm in diam. | Flat colony, smooth, irregular margins | ||
MEA, malt extract agar; PDA, potato dextrose agar.
The diameter of the colony cultured for 7 d on PDA was 24–27 mm, the color of the surface was light gray (Fig. 1A), and the reverse side was light greyish-brown at the center and light gray at the margin (Fig. 1E). The elevation of the colony was flat, and the mycelia grew by digging into the substrate, forming a wrinkled pattern on the plate medium. The colony margins were irregular. The diameter of the colonies cultured for 7 d on MEA was 34–39 mm, and the colonies grew faster than those cultured on PDA. The surface was dark blackish-brown and covered with white fluffy aerial mycelia (Fig. 1B). The reverse was dark brown at the center and light brown at the margin (Fig. 1F). The colony elevation was slightly increased, and the hyphae extended radially to form a smooth margin. Hyaline and globose-tosubglobose chlamydospores were observed at the apex of hyphal growth (Figs. 1I and 1J). The size of the chlamydospores was (9.69–) 13.71 (–16.70) × (7.94–) 11.09 (–14.76) μm in diameter.
Specimen examined. Jinju-si, Gyeongsangnam-do, Korea, 35°12’37.8″N, 128°07’02.7″E, June 5, 2024. D. sinensis, isolated from healthy leaves of S. tuberosum, strain HARI24E001, KACC410788, GenBank Nos. PQ461137 (ITS) and PQ461138 (LSU).
Note. Didymella sinensis was newly reported in 2017 from China, so the epithet ʻsinensis’ was named after its first discovery in China. [16]. In the original description, this species was reported to form leaf spots on cherry leaves [16]; however, it was isolated as an endophyte from healthy tea tree leaves (Camellia sinensis) [17]. In the present study, it was also isolated from healthy potato leaves; therefore, we considered this strain to be a fungal endophyte. Among the morphological characteristics in the original description, it is noteworthy that the colony grew faster on MEA than on PDA, and was covered with fluffy aerial mycelia [16]. The strain HARI24E001 isolated in this study also exhibited these morphological characteristics (Table 1). In the original description, only the teleomorph was identified [16], whereas in this study, conidia from the anamorphic stage were not confirmed. Instead, we confirmed chlamydospores, a general morphological characteristic of the genus Didymella when cultured in vitro [17,18]. The DNA sequence of the ITS region showed 98.09% identity with D. sinensis MK503799.1, and the LSU region showed 99.35% identity with D. sinensis MK503810.1. Strain HARI24E001 formed a monophyletic group with D. sienensis strain MFLUCC 17-1778 and voucher LC8143 in the NJ phylogenetic tree (Fig. 2).
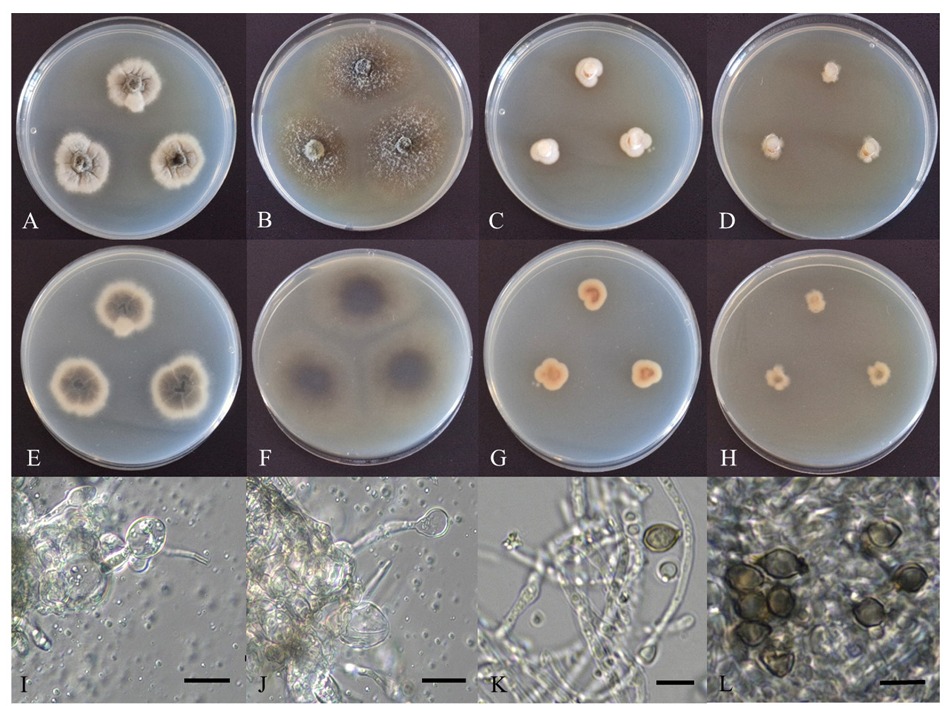
Fig. 1. Cultural characteristics of endophytic fungal strains. Colonies of D. sinensis HARI24E001 grown for 7 d on potato dextrose agar (PDA) (A: surface, E: reverse), malt extract agar (MEA) (B: surface, F: reverse), and chlamydospore (I, J); Colonies of S. oligospora HARI24E013 grown for 7 d on PDA (C: surface, G: reverse), MEA (D: surface, H: reverse), and conidiophore and conidia (K, L) (scale bars: I, J = 20 μm; K, L = 5 μm).
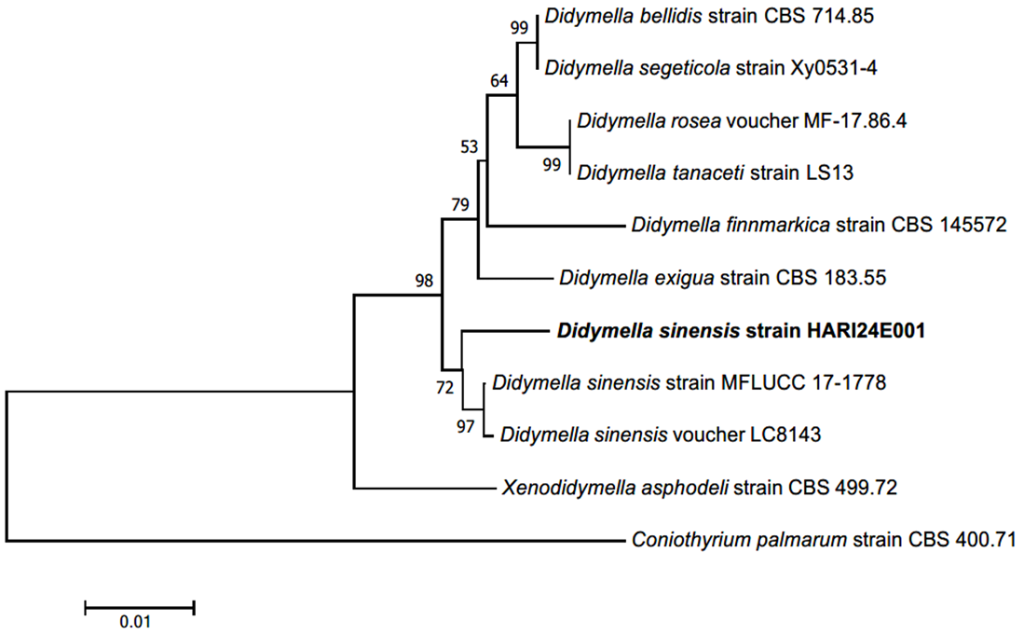
Fig. 2. Neighbor-joining (NJ) phylogenetic tree of based on a concatenated alignment of internal transcribed spacer (ITS) and large subunit (LSU) rDNA sequences. Coniothyrium palmarum was used as an outgroup. Numbers on branches indicate bootstrap values (1,000 replicates). The fungal strain isolated in this study (HARI24E001) is indicated in a bold. Didymella exigua (CBS 183.55) was the type strain.
The diameter of the colonies cultured on PDA for 7 d grew slowly to 14–15 mm. The surface was bright white (Fig. 1C), the reverse was overall beige, and the central part of the colony was reddish-brown (Fig. 1G). The colony was flat on the medium plate, lacked aerial hyphae, and formed a smooth but irregular margin. The diameter of the colonies cultured on MEA for 7 d was 9–11 mm, as they grew slowly. The surface was light beige (Fig. 1D), and the reverse was dark beige; however, a dark brown color was observed in the center, which was presumed to be pigmentation due to conidial maturity (Fig. 1H). The colony elevation was flat and showed highly irregular margins. Conidiophores occurred in the apical or lateral part of the hyphal growth (Fig. 1K). The conidia were ovoid or fusiform with thick spore walls, and a cicatrix (scar) of segmentation from the conidiophores was often observed at the apex and base. Occasionally, rough warts were observed on the surface of the spore walls. The color of the conidia was light yellowish-brown, and the size was (3.92–) 4.58 (–5.52) × (1.91–) 3.18 (–3.88) μm in diameter (Fig. 1L).
Specimen examined. Cheongju-si, Chungcheongbuk-do, Korea, 36°43’34.3″N, 127°27’58.6″E, June 5, 2024. Sagenomella oligospora, isolated from healthy leaves of Solanum tuberosum, HARI24E013, KACC410789, GenBank Nos. PQ461140 (ITS), PQ461139 (LSU), and PQ570856 (TUB).
Note. Sagenomella oligospora was first reported in 1978 as a strain isolated from agricultural soil in the Netherlands, and the genus Sagenomella was separated from the genus Acremonium based on the morphological characteristics of the conidia [19]. It is characterized by the generation of a conidium from the apex of an existing conidium; thus, the conidia appear to be connected in a chain [19]. The conidia observed in the present study formed clusters; however, the chain form was not observed because the conidia were already segmented. Notably, the cicatrix of the connected chain was sometimes observed at the apex and base of the conidia, which protruded into a conical shape. We confirmed the consistency of morphological characteristics such as pigmentation, thick spore walls, and warts on the surface of the spore wall (Table 1). The DNA sequence of the ITS region showed 99.27% identity with S. oligospora MT732892.1, the LSU region showed 100% identity with S. oligospora LC177635.1, and the TUB region showed 98.68% identity with S. oligospora LT634016.1. Strain HARI24E013 formed a monophyletic group with S. oligospora strains CBS 168.74, and CCF 1552 in the NJ phylogenetic tree (Fig. 3).
Through this identification process, we identified two previously unrecorded fungal species in Korea. The identified fungal strains are expected to be useful for potato cultivation and disease control.
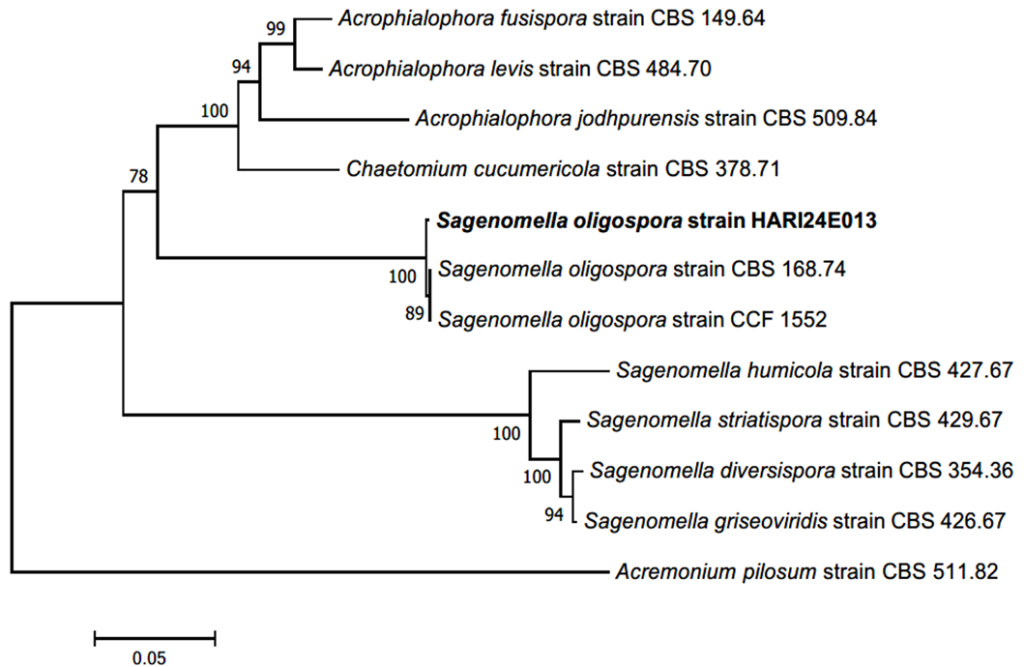
Fig. 3. Neighbor-joining (NJ) phylogenetic tree of based on a concatenated alignment of internal transcribed spacer (ITS), large subunit (LSU), and β-tubulin DNA (TUB) sequences. Acremonium pilosum was used as an outgroup. Numbers on branches indicate bootstrap values (1,000 replicates). The fungal strain isolated in this study (HARI24E013) is indicated in a bold. Sagenomella diversispora (CBS 354.36) was the type strain.
None.
This study was supported by the Rural Development Administration (grant number: PJ017405022024).
1. Carroll G. Fungal endophytes in stems and leaves: from latent pathogen to mutualistic symbiont. Ecology 1988;69:2-9. [DOI]
2. Gao FK, Dai CC, Liu XZ. Mechanisms of fungal endophytes in plant protection against pathogens. Afr J Microbiol Res 2010;4:1346-51.
3. Eo JK, Choi JW, Eom AH. Diversity, distribution, and host plant of endophytic fungi: a focus on Korea. Mycobiology 2022;50:399-407. [DOI]
4. Choi HS, Park H, Eo JK, Eom AH. Unrecorded endophytic fungi isolated from Mnium heterophyllum and Hypnum plumaeforme in Korea: Biscogniauxia petrensis and Cercophora thailandica. Kor J Mycol 2020;48:55-61.
5. Lugtenberg BJ, Caradus JR, Johnson LJ. Fungal endophytes for sustainable crop production. FEMS Microbiol Ecol 2016;92:fiw194. [DOI]
6. Park YE, Cho JH, Im JS, Cho KS, Kim JS, Lee YG, Chang DC, Jin YI, Cheon CK, Jung JC. ‘Dami’, a potato cultivar with few tuber physical defects, high dry matter content, and good taste. Korean J Breed Sci 2019;51:122-7. [DOI]
7. Lee GB, Choi JG, Kwon DH, Yi JY, Lee HT, Jin YI. Effect of spring potato cultivation period on growth, yield and processing quality of autumn potato cultivars. Korean J Plant Res 2024;37:149-60.
8. Zhang L, Xu W, Zhao Z, Long Y, Fan R. Biocontrol potential and growth-promoting effect of endophytic fungus Talaromyces muroii SD1-4 against potato leaf spot disease caused by Alternaria alternata. BMC Microbiol 2024;24:255. [DOI]
9. Lahlali R, Hijri M. Screening, identification and evaluation of potential biocontrol fungal endophytes against Rhizoctonia solani AG3 on potato plants. FEMS Microbiol Lett 2010;311:152-9. [DOI]
10. El-Hasan A, Ngatia G, Link TI, Voegele RT. Isolation, identification, and biocontrol potential of root fungal endophytes associated with solanaceous plants against potato late blight (Phytophthora infestans). Plants 2022;11:1605. [DOI]
11. Park H, Jung CR, Eom AH. Species diversity and antifungal activity of endophytic fungi isolated from Angelica gigas Nakai. Kor J Mycol 2021;49:497-505.
12. Gardes M, Bruns TD. ITS primers with enhanced specificity for basidiomycetes-application to the identification of mycorrhizae and rusts. Mol Ecol 1993;2:113-8. [DOI]
13. Vilgalys R, Hester M. Rapid genetic identification and mapping of enzymatically amplified ribosomal DNA from several Cryptococcus species. J Bacteriol Res 1990;172:4238-46. [DOI]
14. Glass NL, Donaldson GC. Development of primer sets designed for use with the PCR to amplify conserved genes from filamentous ascomycetes. Appl Environ Microbiol 1995;61:1323-30. [DOI]
15. Kumar S, Stecher G, Tamura K. MEGA7: molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol Biol Evol 2016;33:1870-4. [DOI]
16. Chen Q, Hou LW, Duan WJ, Crous PW, Cai L. Didymellaceae revisited. Stud Mycol 2017;87:105-59. [DOI]
17. Wang Y, Tu Y, Chen X, Jiang H, Ren H, Lu Q, Wei C, Lv W. Didymellaceae species associated with tea plant (Camellia sinensis) in China. MycoKeys 2024;105:217-51. [DOI]
18. Chen Q, Jiang JR, Zhang GZ, Cai L, Crous PW. Resolving the Phoma enigma. Stud Mycol 2015;82:137-217. [DOI]
19. Gams W. Connected and disconnected chains of phialoconidia and Sagenomella gen. nov. segregated from Acremonium. Persoonia-Mol Phyl Evol Fungi 1978;10:97-112.