Mohammad Hamizan Azmi1, Seong-Keun Lim1, Ye-Seo Lee1, Jun-Woo Choi1, Min-Gyu Kim1, SeungYeol Lee1,2*, and Hee-Young Jung1,2
1Department of Plant Medicine, Kyungpook National University, Daegu 41566, Korea
2Institute of Plant Medicine, Kyungpook National University, Daegu 41566, Korea
*Correspondence to leesy1123@knu.ac.kr
Korean Journal of Mycology (Kor J Mycol) 2024 December, Volume 52, Issue 4, pages 265-275.
https://doi.org/10.4489/kjm.520405
Received on November 04, 2024, Revised on December 02, 2024, Accepted on December 02, 2024, Published on Dec 30, 2024.
Copyright © The Korean Society of Mycology.
This is an Open Access article which is freely available under the Creative Commons Attribution-NonCommercial 4.0 International License (CC BY-NC) (https://creativecommons.org/licenses/by-nc/4.0/).
During an exploration of fungal diversity in Korean soil, this study isolated and identified two unrecorded fungal strains, designated KNU-HP-1804 and KNUF-20-007. Morphological and cultural features were examined for initial classification. Cultural and morphological characteristics of strains KNU-HP-1804 and KNUF-20-007 were matched with Clonostachys eriocamporesii CBS 647.91 and Sporothrix euskadiensis CBS 122138T, respectively. To further pinpoint their identity and evolutionary relationships, molecular phylogenetic analyses were conducted using the internal transcribed spacer (ITS) region, translation elongation factor 1-alpha (TEF1), beta-tubulin (TUB2), and calmodulin (CAL). The neighbor-joining (NJ) phylogenetic tree using the concatenated sequences, and the cultural and morphological observations revealed KNU-HP-1804 as C. eriocamporesii, while KNUF-20-007 was identified as S. euskadiensis. To the best of our knowledge, this is the first report on C. eriocamporesii and S. euskadiensis in Korea.
Clonostachys eriocamporesii, Cultural characteristics, Morphology, Phylogenetic analyses, Sporothrix euskadiensis
Fungi play an important ecological role as decomposers of organic matter. Fungi produce a wide range of enzymes that help regulate the balance of soil nutrients, greatly influencing plant productivity and diversity, and they also act as predators, pathogens, and parasites of many organisms [1,2]. The fungi in genus Clonostachys, belongs to the class Sordariomycetes, order Hypocreales, family Bionectriaceae, with 68 species reported worldwide [3]. The family Bionectriaceae is characterized by its herbicolous, corticolous, lichenicolous, fungicolous, and coprophilous nature [4]. The genus Clonostachys has recently been found in the bark and decaying leaves of dead trees, other fungi, nematodes, and insects, and comprises soil-borne species, mycoparasites, endophytes, epiphytes, and saprotrophs [4,5]. The Clonostachys species identified in Korea currently comprises three recognized species: C. rosea, C. divergens, and C. farisona [6–8]. The genus Sporothrix also belongs to the class Sordariomycetes, but is in the order Ophiostomatales, and family Ophiostomataceae, with 56 species reported worldwide [9,10]. In 2016, a study confirmed that most species with Sporothrix-like asexual morphs do not constitute a monophyletic lineage in the genus Ophiostoma, and was reclassified into the genus Sporothrix [11]. The genus Sporothrix is widely distributed in different climatic zones of the world and has been reported from a variety of habitats associated with forest trees, soil, bark, beetles and mites [12]. In addition, the genus Sporothrix comprises soil-borne, saprophytic, endophytic, and phytophytic bacteria and includes S. brasiliensis, S. chilensis, S. globose, S. luriei, and S. schenskii, which have been reported as toxic pathogens of livestock [13–15]. Currently, there are 8 reported species in Korea under the genus Sporothrix [16–18].
The objective of this research was to isolate fungi from soil samples collected from Korea and to identify the isolated fungal species through morphological and molecular biological characteristics.
The fungal isolates used in this study were present in soil samples collected from Gunwi-gun, Gyeongbuk province (36°08’56.0″N 128°35’46.3″E) and Yangpyeong-gun, Gyeonggi province (37°28’03.7″N 127° 32’30.3″E) in Korea. The isolation of fungi from the soil samples followed by the serial dilution technique was described in a prior study [19]. Colonies displaying signs of germination were then transferred to new potato dextrose agar (PDA; Difco, Detroit, MI, USA) plates at a temperature of 25℃. The fungal strains KNU-HP-1804 and KNUF-20-007 were chosen for additional molecular analyses, as well as cultural and morphological evaluations. The stock culture for strain KNU-HP-1804 (NIBRFGC000502238) and strain KNUF-20-007 (NIBRFGC000507843) have been deposited at the National Institute of Biological Resources (NIBR) as a metabolically inactive culture.
Different media were used to observe the culturological and morphological characteristics of the two strains. The strain KNU-HP-1804 was cultured for 7 days on PDA, oatmeal agar (OA; Difco, Detroit, MI, USA), and synthetic nutrient deficient agar (SNA; Nirenberg, 1976); while strain KNUF-20-007 was cultured for 8 days on PDA, OA, and 2% malt extract agar (MEA; Difco, Detroit, MI, USA), with the growth, color, morphology, and texture of the colonies formed on were observed [3,20]. Morphological features were observed and recorded using a light microscope (BX-50, Olympus, Tokyo, Japan).
The total genomic DNA was extracted from the strains KNU-HP-1804 and KNUF-20-007 using the HiGene™ Genomic DNA Prep Kit (Biofact, Daejeon, Korea) for molecular identification. KNU-HP-1804 was analyzed for the internal transcribed spacer (ITS) region, translation elongation factor 1-alpha (TEF1), and beta-tubulin (TUB2) sequence fragments, while the strain KNUF-20-007 was analyzed for the ITS region, TUB2, and calmodulin (CAL) sequence fragments. The primers used to amplify the molecular phylogenetic markers were ITS1F/ITS4 for ITS, EF1-983F/EF1-2218R for TEF1, CL2F/CL2R for CAL, and T1/T22 and Bt2a/Bt2b for TUB2 [21–26]. The confirmation of amplification was conducted through electrophoresis utilizing 1.0% HP Agarose gels (BIOPURE, Cambridge, USA). The amplified products underwent purification via ExoSAP-IT (Thermo Fisher Scientific, Waltham, MA, USA) before being sent to Macrogen (Seoul, Korea) for sequencing.
The sequences obtained were analyzed for similarity utilizing the Basic Local Alignment Search Tool (BLAST) within the National Center for Biotechnology Information (NCBI) database (Table 1). Phylogenetic trees were constructed from the combined sequences of the ITS regions, TEF1, TUB2, and CAL employing the neighbor-joining (NJ) method in Molecular Evolutionary Genetics Analysis (MEGA) version 11.0 [27,28]. The evolutionary distance matrices for the NJ analysis were calculated in accordance with Kimura’s two-parameter model, with bootstrap values derived from 1,000 replications [29].
Table 1. List of species and their GenBank accession numbers used for the phylogenetic analyses in this study
| Species name | Strain number | GenBank accession number | |||
|---|---|---|---|---|---|
| ITS | TEF1 | TUB2 | CAL | ||
| Clonostachys fujianensis | CBS 12747T | OQ910620 | OQ982655 | OQ944632 | – |
| Clonostachys obovatispora | CBS 118752T | OQ910649 | OQ982680 | OQ944661 | – |
| Clonostachys epichloe | CBS 101037T | OQ910581 | OQ982618 | OQ944593 | – |
| Clonostachys miodochialis | CBS 997.69T | OQ910646 | OQ982677 | OQ944658 | – |
| Clonostachys divergens | JW190011 | OQ910578 | OQ982615 | OQ944590 | – |
| Clonostachys divergens | CBS 967.73BT | OQ910575 | OQ982612 | OQ944587 | – |
| Clonostachys samuelsii | CBS 699.97T | OQ910812 | OQ982832 | OQ944822 | – |
| Clonostachys samuelsii | CBS 701.97 | OQ910814 | OQ982834 | OQ944824 | – |
| Clonostachys rogersoniana | CBS 920.97T | OQ910711 | OQ982740 | OQ944723 | – |
| Clonostachys rogersoniana | CBS 668.70 | OQ910710 | OQ982739 | OQ944722 | – |
| Clonostachys penicillata | CBS 729.87T | OQ910654 | OQ982685 | OQ944666 | – |
| Clonostachys penicillate | CBS 148211 | OQ910652 | OQ982683 | OQ944664 | – |
| Clonostachys hongkongensis | CBS 118291T | OQ910630 | OQ982663 | OQ944642 | – |
| Clonostachys eriocamporesii | CBS 647.91 | OQ910582 | OQ982619 | OQ944594 | – |
| Clonostachys eriocamporesii | KNU-HP-1804 | PQ269427 | PQ276740 | PQ276750 | – |
| Fusarium acutatum | CBS 402.97T | NR_111142 | MT011051 | MT010989 | – |
| Sporothrix cf. abietina | CMW40454 | MW581512 | – | MW579723 | MW579751 |
| Sporothrix abietina | CBS 125.89T | AF484453 | – | KX590755 | JQ511966 |
| Sporothrix lunata | CMW10563T | AY280485 | – | AY280466 | JQ511970 |
| Sporothrix prolifera | KFL99WRJSI | MH283139 | – | MH283355 | MH283520 |
| Sporothrix prolifera | CBS 251.88T | KX590829 | – | KX590770 | KX590797 |
| Sporothrix cantabriensis | CMW39766T | KF951554 | – | KF951544 | KF951540 |
| Sporothrix cantabriensis | CMW39767 | KF951555 | – | KF951545 | KF951541 |
| Sporothrix fusiformis | CMW9968T | AY280481 | – | AY280461 | JQ511967 |
| Sporothrix fusiformis | CMW7131 | AY280497 | – | AY280464 | JQ511971 |
| Sporothrix gossypina | ATCC 18999T | KX590819 | – | KX590761 | KX590789 |
| Sporothrix curviconia | CBS 541.84 | KX590836 | – | KX590777 | JQ511968 |
| Sporothrix rossii | CBS 116.78T | NR_147597 | – | KX590754 | JQ511972 |
| Sporothrix euskadiensis | CBS 122138T | DQ674369 | – | EF396344 | JQ438830 |
| Sporothrix euskadiensis | KNUF-20-007 | PQ269428 | – | PQ276751 | PQ276741 |
| Ophiostoma noisomeae | CBS 141065 | KU639631 | – | KU639628 | KX590792 |
ITS: Internal transcribed spacer regions; TEF1: translation elongation factor-1ɑ; TUB2: beta-tubulin; CAL: calmodulin. TType strain; The strain used in this research is highlighted in bold.
The colony grown on PDA at 25℃ for 7 days was observed to be growing 31.7–34.5 mm in diameter. The obverse side appears to be white with cottony aerial hyphae. The reverse side was observed to be yellowish white (Fig. 1A). Grown on OA at 25℃ for 7 days, the colony was observed to be 37.7–38.2 mm in diameter. On the obverse side was observed to be white and cottony, forming a crateriform in the center. The reverse side showed the same white color as the obverse side (Fig. 1B). Grown on SNA at 25℃ for 7 days, the colony was observed to be 24.7–25.5 mm, appeared to be yellowish on both obverse and reverse sides, and were observed to be rhizoid (Fig. 1C). The conidiophores, which produces conidia, were transparent, smooth-surfaced, monomorphic and sporodochial. The phialides were upright, cylindrical, tapers towards the apex and narrowly flask shaped. Phialides measured at 7.9–17.5 μm (avg. 10.9 μm, n = 20) (Fig. 1D). When incubated for one month on PDA and SNA, dark green, long, curled stuppeus conidia were observed. White pustules were also observed to first form on aerial hyphae, followed by light-green sporodochia which turns to dark-green sporodochia over time (Figs. 1E and F). Conidia produced at the tip of phialides were transparent unicellular with a thin, smooth surface, fusiform with a pointed tip, and were layered and chained like scales, measuring 4.5–7.0 × 1.7–3.4 μm (Figs. 1G and H). A comparison of the morphological features between strain KNU-HP-1804 and Clonostachys eriocamporesii CBS 647.91 was made in Table 2. Based on the table, the strain KNU-HP-1804 was indicated to be closely related to C. eriocamporesii CBS 647.91 [3].
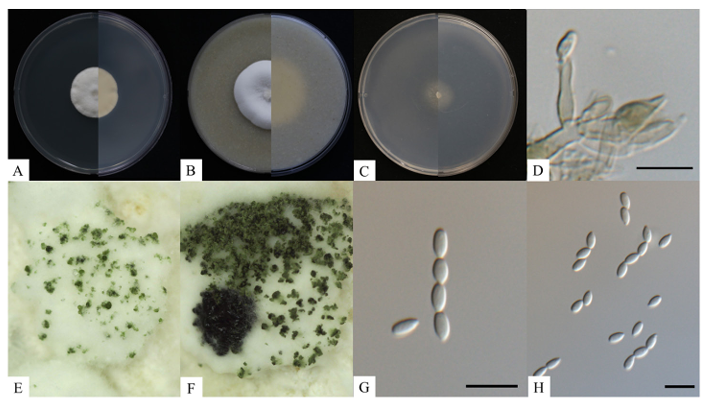
Fig. 1. Cultural and morphological characteristics of Clonostachys eriocamporesii KNU-HP-1804. Cultures were grown at 25℃ for 7 days. A–C: Front and reverse colony on potato dextrose agar (PDA), oatmeal agar (OA), synthetic nutrient deficient agar (SNA), respectively; D: Conidiophores; E: Sporodochia after 1 month; F: Sporodochia after 2 months; G, H: Conidia. Scale bars = 10 μm.
Table 2. Morphological characteristics of KNU-HP-1804 and comparisons between Clonostachys eriocamporesii CBS 647.91
| Characteristics | Clonostachys eriocamporesii KNU-HP-1804a | Clonostachys eriocamporesii CBS 647.91b | |
|---|---|---|---|
| Colony on PDA | Size | 27 mm | 32–41 mm |
| Color | White, reverse concolorous | White, reverse concolorous | |
| Shape | Margin entire, aerial mycelium moderate, felty to cottony | Margin entire, aerial mycelium moderate, felty to cottony | |
| Colony on OA | Size | 37–38 mm | 30–45 mm |
| Color | White, reverse concolorous | White, reverse concolorous | |
| Shape | Margin entire, aerial mycelium moderate, felty to cottony | Margin entire, aerial mycelium moderate, felty to cottony | |
| Colony on SNA | Size | 23 mm | 27–32 mm |
| Color | Yellowish-white, reverse concolorous | White, reverse concolorous | |
| Shape | Rhizoid | Margin entire, aerial mycelium moderate in center, floccose | |
| Sporodochia | Shape | White pustules, light green to dark green | White pustules, dark green |
| Conidiophores | Size | 7.9–17.5 μm | 9.5–17.5 μm |
| Shape | Monomorphic, sporodochial | Monomorphic, sporodochial | |
| Conidia | Size | 4.5–7.0 × 1.7–3.4 μm | 5.9–8.6 × 2.7–3.8 μm |
| Shape | Hyaline, narrowly clavate, arranged in chain | Aseptate, greenish hyaline, ellipsoidal, narrowly clavate, arranged in chain |
PDA: potato dextrose agar; OA: oatmeal agar; SNA: synthetic nutrient deficient agar.
aFungal strain used in this study; b Source of description [3].
To genetically identify fungal strain KNU-HP-1804, nucleotide sequences of ITS regions, TEF1, and TUB2 were acquired with the length 485, 724, and 1031 bp, respectively. The nucleotide sequences were registered to GenBank under the accession numbers PQ269427, PQ276740, PQ276750 for the ITS regions, TEF1, and TUB2, respectively. The ITS regions showed the highest similarity of 99.8% with C. eriocamporesii CBS 647.91, when compared with C. epichloe CBS 101037T at a similarity of 93.2%. The TEF1 sequence showed a very high similarity of 99.0% with C. eriocamporesii CBS 647.91 compared to a similarity of 97.9% with C. miodochialis CBS 997.69T while for TUB2, KNU-HP-1804 showed the highest similarity of 100% with C. eriocamporesii CBS 647.91. The NJ phylogenetic tree constructed based on the concatenated sequences of the ITS region, TEF1, and TUB2 (Fig. 2), showed that strain KNUHP-1804 formed the same cluster as the reported C. eriocamporesii and was identified as C. eriocamporesii based on culture, morphological, and phylogenetic analyses. This report on C. eriocamporesii contributes to the existing knowledge of Clonostachys species, bringing the number of recorded species to 4 in Korea [6–8]. Species from the genus Clonostachys were reported to be entomopathogenic, with C. eriocamporesii reported to cause fast and high mortality in Aedes aegypti larvae and were found on invasive spotted lanternflies [30,31]. There were also several reports of C. rosea as opportunistic phytopathogens and naturally occurring entomopathogen or a promising control agent [8,30]. The mycorrhizal and saprophytic species of the genus Clonostachys are capable of destroying plant pathogens by secreting chitinolytic enzymes, proteolytic enzymes, and several antibiotics, and possess induced resistance that can protect plants from pathogen invasion [32,33]. It has been reported that C. miodochialis also produces metabolites that may be antagonistic to F. acuminatum and may be involved in degrading the cell wall without physical contact [34]. As such, biological control using the genus Clonostachys has been actively pursued, and further research on C. eriocamporesii is warranted.
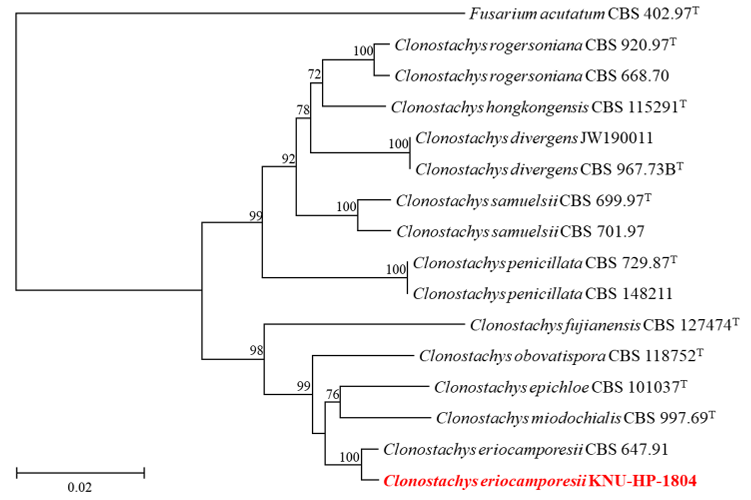
Fig. 2. Neighbor-joining phylogenetic analyses of KNU-HP-1804 based on concatenated sequence data of internal transcribed spacer (ITS) regions, translation elongation factor 1-alpha (TEF1) and betatubulin (TUB2) showing the phylogenetic position of the closest species in the genus Clonostachys. Bootstrap values greater than 70% (percentage of 1,000 replications) are shown at branching points. The strain isolated in this study was highlighted in bold and red, and bootstrap values were obtained from 1,000 replicates. Fusarium acutatum CBS 402.97T was used as an outgroup. Bar = 0.02 substitutions per nucleotide position
The colony grown on PDA for 8 days at 25℃ was observed to be growing 31.7–34.5 mm in diameter. On the obverse side, the colony appears yellowish-white with aerial hyphae that were filamentous (Fig. 3A). Grown on OA for 8 days at 25℃, the colony was observed to be 22.3–23.4 mm in diameter. The colony appears white and filiform on the obverse side, and pale white on the reverse side (Fig. 3B). Colony grown on MEA for 8 days at 25℃ measured to be 18.7–19.0 mm in diameter. The colony was filamentous, with both obverse and reverse sides being yellowish-white in color (Fig. 3C). The conidiophores were transparent, smooth-surfaced, with conidia hanging from the apex of an elongated cylindrical shape, and measured 6.1–23.4 μm (avg. 13.7 μm, n = 20) in length (Fig. 3D). The transparent unicellular conidia were clavate and cylindrical in shape, and measured to be 3.1–5.4 × 1.4–2.3 μm (n = 50) (Figs. 3E and F). A comparison of the morphological features between strain KNUF-20-007 and S. euskadiensis CBS 122138T was made in Table 2. Based on the table, the strain KNUF-20-007 was indicated to be closely related to S. euskadiensis CBS 122138T [11].
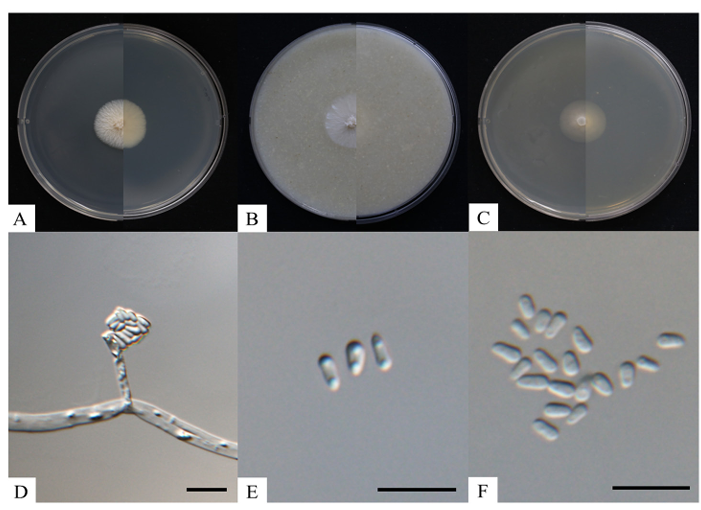
Fig. 3. Cultural and morphological characteristics of Sporothrix euskadiensis KNUF-20-007. Cultures were grown at 25℃ for 8 days. A-C: Front and reverse colony on potato dextrose agar (PDA), oatmeal agar (OA) and 2% malt extract agar (MEA), respectively; D: Sporothrix-like conidiophore; E, F: Conidia. Scale bars = 10 μm.
Table 3. Morphological characteristics of KNUF-20-007 and comparisons between Sporothrix euskadiensis CBS 122138T
| Characteristics | Sporothrix euskadiensis KNUF-20-007a | Sporothrix euskadiensis CBS 122138Tb | |
|---|---|---|---|
| Colony on PDA | Size | 31.7–34.5 mm | N/A |
| Shape and color | Yellowish-white, reverse concolorous, filamentous, hyphae aerial | N/A | |
| Colony on OA | Size | 22.3–23.4 mm | N/A |
| Shape and color | White, reverse side concolorous, filiform | N/A | |
| Colony on MEA | Size | 18.7–19.0 mm | 14.4 mm |
| Shape and color | Yellowish-white, filamentous, entire margin | Yellowish-white, entire margin | |
| Conidiophores | Size | 6.1–23.4 μm | 10.2–10.8 μm |
| Shape | Transparent, smooth-surfaced, conidia hanging from apex of an elongated cylindrical shape | N/A | |
| Conidia | Size | 3.1–5.4 × 1.4–2.3 μm | 2.2–3.0 × 1.2–1.8 μm |
| Shape | Clavate, cylindrical | Clavate |
PDA: potato dextrose agar; OA: oatmeal agar; MEA: malt extract agar.
aFungal strain used in this study; b Source of description [11]; TType strain; N/A: not available.
To genetically identify the fungal strain KNUF-20-007, nucleotide sequences of ITS regions, CAL, and TUB2 were acquired with the length 611, 521, and 256 bp, respectively. The nucleotide sequences were registered to GenBank under the accession numbers PQ269428, PQ276751, PQ276741 for the ITS regions, TUB2, and CAL, respectively. The ITS regions revealed the highest similarity of 99.8% with S. euskadiensis CBS 122138T, compared to a similarity of 99.6% with S. cf. abietina CMW40454, and 99.5% with S. rossii CBS 116.78T. Furthermore, the CAL sequences showed a very high similarity of 99.5% with S. euskadiensis CBS 122138T, compared with a similarity of 97.8% with S. cf. abietina CMW40454. Based on the TUB2 sequences, strain KNUF-20-007 showed a 100% similarity with S. euskadiensis CBS 112138T. A NJ phylogenetic tree was constructed using the concatenated ITS region, CAL and TUB2 sequences (Fig. 4). The NJ phylogenetic tree indicated that strain KNUF-20-007 was clustered with S. euskadiensis CBS 112138T. Based on the morphological, cultural, and phylogenetic analysis, the strain KNUF-20-007 was identified as S. euskadiensis. Including this report on S. euskadiensis, currently there are 9 reported species of Sporothrix in Korea [16]. The genus Sporothrix is mostly a soil-borne species native to tropical regions, but it has been reported to be isolated from insects such as beetles and mites that live in the bark of trees. While there has been no reports of S. euskadiensis being naturally pathogenic or an entomopathogenic fungi; S. schenckii, which is reported to be an endophytic pathogen, has been isolated from decaying plants, rose bushes, and sphagnum moss, causing a chronic fungal infection known as sporotrichosis [15]. However, as many species of the genus Sporothrix are reported to be toxic pathogens of livestock, it is necessary to obtain additional isolates and conduct pathogenicity tests to characterize S. euskadiensis. This study contributed to increasing the diversity of endemic fungal species by discovering C. eriocamporesii and S. euskadiensis, which to the best of our knowledge, have not been reported in Korea. It also contributed to discovering their utilization value through further research and increasing the possibility of their application in various industrial fields.
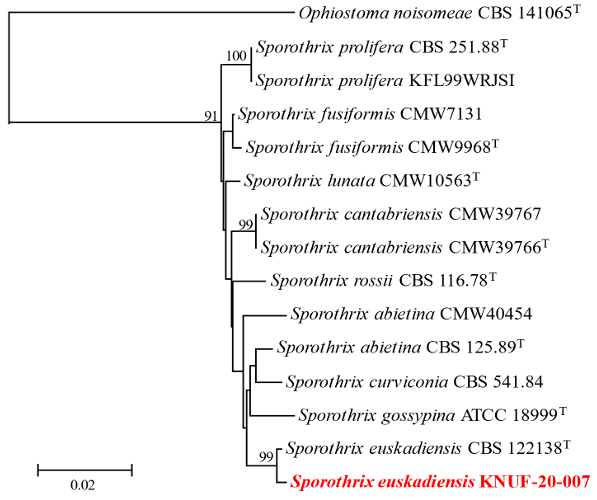
Fig. 4. Neighbor-joining phylogenetic analyses of KNUF-20-007 based on concatenated sequence data of internal transcribed spacer (ITS) regions, calmodulin (CAL) and beta-tubulin (TUB2) showing the phylogenetic position of the closest species in the genus Sporothrix. Bootstrap values greater than 90% (percentage of 1,000 replications) are shown at branching points. The strain isolated in this study was highlighted in bold and red, and bootstrap values were obtained from 1,000 replicates. Ophiostoma noisomeae CBS 141065T was used as an outgroup. Bar = 0.02 substitutions per nucleotide position.
No conflict of interest is declared by the author.
This research was supported by a grant from the National Institute of Biological Resources (NIBR), funded by the Ministry of Environment (MOE) of the Republic of Korea (NIBR201801105 and NIBR202002104).
1. Frąc M, Hannula SE, Bełka M, Jędryczka M. Fungal biodiversity and their role in soil health. Front Microbiol 2018;9:707. [DOI]
2. Gebrie SA. Biotrophic fungi infection and plant defense mechanism. J Plant Pathol Microbiol 2016;7:378. [DOI]
3. Zhao L, Groenewald JZ, Hernández-Restrepo M, Schroers HJ, Crous PW. Revising Clonostachys and allied genera in Bionectriaceae. Stud Mycol 2023;105:205-66. [DOI]
4. Schroers HJ. A monograph of Bionectria (Ascomycota, Hypocreales, Bionectriaceae) and its Clonostachys anamorphs. Stud Mycol 2001;46:1-11.
5. Moreira GM, Abreu LM, Carvalho VG, Schroers HJ, Pfenning LH. Multilocus phylogeny of Clonostachys subgenus Bionectria from Brazil and description of Clonostachys chloroleuca sp. nov. Mycol Prog 2016;15:1031-9. [DOI]
6. Romón P, de Beer ZW, Zhou X, Duong TA, Wingfield BD, Wingfield MJ. Multigene phylogenies of Ophiostomataceae associated with Monterey pine bark beetles in Spain reveal three new fungal species. Mycologia 2014;106:119-32. [DOI]
7. You YH, Park JM, Park JH, Kim JG. Diversity of endophytic fungi associated with the roots of four aquatic plants inhabiting two wetlands in Korea. Mycobiology 2015;43:231-8. [DOI]
8. Lee SA, Kang MJ, Kim TD, Park EJ. First report of Clonostachys rosea causing root rot of Gastrodia elata in Korea. Plant Dis 2020;104:3069. [DOI]
9. Wang HM, Wang Z, Liu F, Wu CX, Zhang SF, Kong XB, Decock C, Lu Q, Zhang Z. Differential patterns of ophiostomatoid fungal communities associated with three sympatric Tomicus species infesting pines in south-western China, with a description of four new species. MycoKeys 2019;50:93-133. [DOI]
10. Musvuugwa T, de Beer ZW, Dreyer LL, Duong T, Marincowitz S, Oberlander KC, Roets F. New ophiostomatoid fungi from wounds on storm-damaged trees in Afromontane forests of the Cape Floristic Region. Mycol Prog 2020;19:81-95. [DOI]
11. de Beer ZW, Duong TA, Wingfield MJ. The divorce of Sporothrix and Ophiostoma: solution to a problematic relationship. Stud Mycol 2016;83:165-91. [DOI]
12. de Meyer EM, de Beer ZW, Summerbell RC, Moharram A, de Hoog GS, Vismer HF, Wingfield MJ. Taxonomy and phylogeny of new wood-and soil-inhabiting Sporothrix species in the Ophiostoma stenoceras-Sporothrix schenckii complex. Mycologia 2008;100:647-61. [DOI]
13. López-Romero E, Reyes-Montes MdR, Pérez-Torres A, Ruiz-Baca E, Villagómez-Castro JC, Mora-Montes HM, Flores-Carreón A, Toriello C. Sporothrix schenckii complex and sporotrichosis, an emerging health problem. Future Microbiol 2011;6:85-102. [DOI]
14. Zhang Y, Hagen F, Stielow B, Rodrigues AM, Samerpitak K, Zhou X, Feng P, Yang L, Chen M, Deng S. Phylogeography and evolutionary patterns in Sporothrix spanning more than 14 000 human and animal case reports. Persoonia 2015;35:1-20. [DOI]
15. Ramírez-Soto MC, Aguilar-Ancori EG, Tirado-Sánchez A, Bonifaz A. Ecological determinants of sporotrichosis etiological agents. J Fungi 2018;4:95. [DOI]
16. Phaund W, Somaly U, Das K, Lee SY, Jung HY. Clonostachys divergens and Chrysosporium merdarium: two new records from soil in Korea. Kor J Mycol 2023;51:91-100.
17. Park SK, Lee SY, Back CG, Kang IK, Ten L, Lee HB, Jung HY. Sporothrix stylites: a new record from field soil in Korea. Kor J Mycol 2017;45:224-8.
18. Adhikari M, Kim SW, Kim HS, Lee HB, Lee YS. Sixteen new records of ascomycetes from crop field soil in Korea. Kor J Mycol 2016;44:271-88. [DOI]
19. Das K, You YH, Lee SY, Jung HY. A new species of Thelonectria and a new record of Cephalotrichum hinnuleum from Gunwi and Ulleungdo in Korea. Mycobiology 2020;48:341-50. [DOI]
20. Ostafińska A, Jankowiak R, Bilański P, Solheim H, Wingfield MJ. Six new species of Sporothrix from hardwood trees in Poland. MycoKeys 2021;82:1-32. [DOI]
21. White TJ, Bruns TD, Lee SB, Taylor JW. Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics. In: Innis MA, Gelfand DH, Sninsky JJ, editors. PCR protocols: a guide to methods and applications. San Diego: Academic Press; 1990. p. 315-22. [DOI]
22. Gardes M, Bruns TD. ITS primers with enhanced specificity for basidiomycetes‐application to the identification of mycorrhizae and rusts. Mol Ecol 1993;2:113-8. [DOI]
23. Rehner SA, Buckley E. A Beauveria phylogeny inferred from nuclear ITS and EF1-α sequences: evidence for cryptic diversification and links to Cordyceps teleomorphs. Mycologia 2005;97:84-98. [DOI]
24. Duong TA, De Beer ZW, Wingfield BD, Wingfield MJ. Phylogeny and taxonomy of species in the Grosmannia serpens complex. Mycologia 2012;104:715-32. [DOI]
25. O’Donnell K, Cigelnik E. Two divergent intragenomic rDNA ITS2 types within a monophyletic lineage of the fungus Fusarium are nonorthologous. Mol Phylogenet Evol 1997;7:103-16. [DOI]
26. Glass NL, Donaldson GC. Development of primer sets designed for use with the PCR to amplify conserved genes from filamentous ascomycetes. Appl Environ Microbiol 1995;61:1323-30. [DOI]
27. Saitou N, Nei M. The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol Biol Evol 1987;4:406-25.
28. Tamura K, Stecher G, Kumar S. MEGA11: molecular evolutionary genetics analysis version 11. Mol Biol Evol 2021;38:3022-7. [DOI]
29. Kimura M. A simple method for estimating evolutionary rates of base substitutions through comparative studies of nucleotide sequences. J Mol Evol 1980;16:111-20. [DOI]
30. Rodrigues J, Rocha LFN, Martinez JM, Montalva C, Humber RA, Luz C. Clonostachys spp., natural mosquito antagonists, and their prospects for biological control of Aedes aegypti. Parasitol Res 2022;121:2979-84. [DOI]
31. Hajek AE, Everest TA, Clifton EH. Accumulation of fungal pathogens infecting the invasive spotted lanternfly, Lycorma delicatula. Insects 2023;14:912. [DOI]
32. Mukherjee PK, Horwitz BA, Herrera-Estrella A, Schmoll M, Kenerley CM. Trichoderma research in the genome era. Annu Rev Phytopathol 2013;51:105-29. [DOI]
33. Hermosa R, Viterbo A, Chet I, Monte E. Plant-beneficial effects of Trichoderma and of its genes. Microbiology 2012;158:17-25. [DOI]
34. Abdellatif L, Fernandez MR, Lokuruge P. Mode of action of potential biocontrol agents against Fusarium species and Cochliobolus sativus. Mycologia 2022;114:476-86. [DOI]