Seung-Yoon Oh* and Yunhyeok Jang
Department of Biology and Chemistry, Changwon National University, Changwon 51140, Korea
*Correspondence to syoh@changwon.ac.kr
Korean Journal of Mycology (Kor J Mycol) 2024 December, Volume 52, Issue 4, pages 413-417.
https://doi.org/10.4489/kjm.520419
Received on December 21, 2024, Revised on December 23, 2024, Accepted on December 24, 2024, Published on Dec 30, 2024.
Copyright © The Korean Society of Mycology.
This is an Open Access article which is freely available under the Creative Commons Attribution-NonCommercial 4.0 International License (CC BY-NC) (https://creativecommons.org/licenses/by-nc/4.0/).
Metarhizium, a genus of entomopathogenic fungi distributed globally, infects a wide range of insect hosts. To date, five species have been documented in South Korea. During a survey of cicada (Cryptotympana atrata)-associated fungal diversity, we discovered an unexpected case of fungal infection. Morphological examination and molecular phylogenetic analysis identified the isolated fungal strain as Metarhizium viridulum, a species not previously reported in South Korea. This study presents the detailed morphological characteristics and phylogenetic position of the isolated M. viridulum strain, expanding our understanding of Metarhizium diversity in South Korea.
Cicada, Entomopathogenic fungi, Fungi, Metarhizium, Unrecorded species
The genus Metarhizium Sorokīn 1879, a group of entomopathogenic fungi with a global distribution, comprises approximately 80 species that are adapted to diverse ecosystems [1]. Metarhizium is widely recognized as a biocontrol agent against insect pests, and offers an environmentally friendly alternative to chemical pesticides [2]. Although traditionally considered host are generalists with a cosmopolitan distribution, recent morphological and molecular analyses have suggested that these fungi are more host- or region-specific than previously thought [1,3]. These findings highlight the need for further studies to clarify their precise distribution and diversity. In South Korea, five Metarhizium species (M. anisopliae, M. guizhouense, M. marquandii, M. pemphigi, and M. rileyi) have been recorded [4], underscoring the potential unexplored diversity harbored in South Korea. In a recent survey of cicada-associated fungal communities, Metarhizium strains were isolated from cicadas (Cryptotympana atrata). In this study, we report the isolation of Metarhizium viridulum for the first time in South Korea, based on morphological and molecular analyses.
Cicada cadavers were collected from Miryang, South Korea (35°21’13.97″ N, 128°45’39.68″ E), in August 2023. Fungal strains were isolated from cadavers on potato dextrose agar (PDA; Difco, Becton Dickinson, MD, USA) at 25℃ and subcultured until pure cultures were obtained. All strains were deposited in the herbarium of Changwon National University (CWNU). For molecular analysis, genomic DNA was extracted from the strains using the AccuPrep Genomic DNA Extraction Kit (Bioneer, Daejeon, Korea). PCR amplification was performed using a SimpliAmp™ Thermal Cycler (Applied Biosystems, Waltham, MA, USA) for the nuclear ribosomal large subunit (LSU) region with LR0R [5] and LR5 primers [6]. Elongation factor 1-alpha (EF1a) was amplified using EF1-983F and EF1-2218R primers [7], following previously described protocols [8]. The PCR results were verified using 1% agarose gel electrophoresis, and the PCR products were sequenced using an ABI 3730xl Sequencer (Macrogen, Seoul, South Korea). Sequences were checked and assembled using MEGA5 [9] and aligned locus-specifically with reference sequences using MAFFT v.7 [10]. Because the M. viridulum type strain sequence is unavailable [11], the sequence of the authentic strain BCC36261, which has been morphologically and molecularly characterized [1], was used for molecular analysis. Maximum-likelihood (ML) phylogenetic analysis was performed with concatenated LSU and EF1a sequences using RAxML v.8 [12] with the GTRGAMMA model and 1000 bootstrap replicates. All sequences from this study have been deposited in GenBank (accession number PQ790513-14). Metarhizium strains CSC23C0001 and CSC23C0002 clustered monophyletically with the reference strain BCC36261 of M. viridulum, with 96% bootstrap support (Fig. 1). In both strains, pairwise comparisons with strain BCC36261 revealed sequence similarities of 100% and 99.77% in the LSU and EF1a region, respectively.
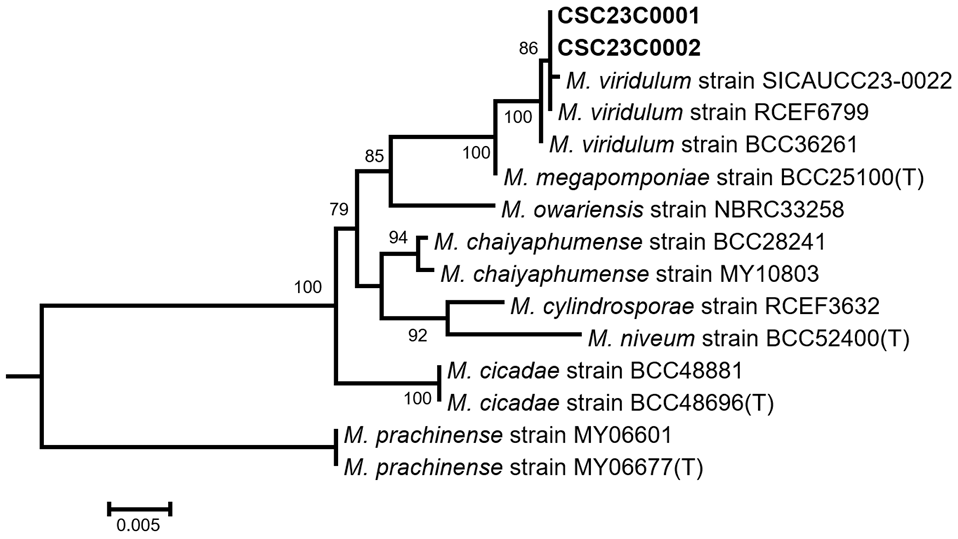
Fig. 1. Maximum-likelihood phylogenetic tree based on concatenated the large subunit ribosomal DNA (LSU) and elongation factor 1-alpha (EF1a) sequences. Bootstrap support values of > 70 are shown. The strains analyzed in this study are in boldface. The scale bar represents the number of substitutions per site.
Cultural characteristics were assessed using three media types: malt extract agar (MEA; Oxoid, Hampshire, UK), oatmeal agar (OA; Difco, Becton Dickinson, MD, USA), and PDA. The micromorphological characteristics were observed using an Eclipse Ni light microscope (Nikon, Tokyo, Japan).
Metarhizium viridulum (Tzean et. al.) B. Huang & Z.Z. Li, Mycosystema 23: 36. 2004
Colony diameter at 25℃ (in mm): After 14 d, MEA: 30–35; OA: 27–31; PDA: 23–26. After 28 d, MEA: 50–75; OA: 75–85; PDA: 40–50 (Fig. 2).
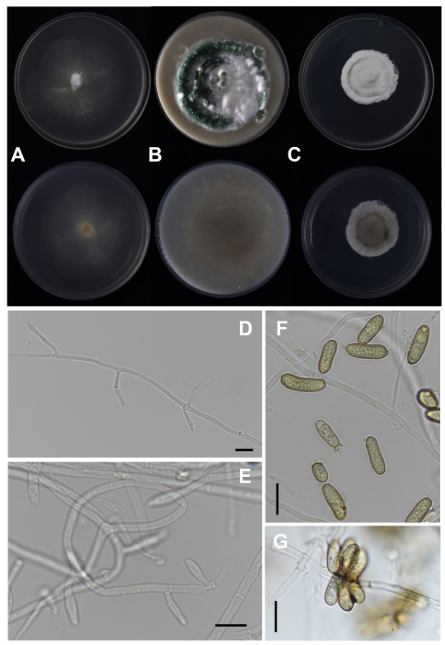
Fig. 2. Metarhizium viridulum strain CSC23C0001 in 28-day-old cultures grown at 25℃. (A–C) Colonies on 90 mm plate of malt extract agar (MEA), oatmeal agar (OA), and potato dextrose agar (PDA), shown from left to right (top: obverse view, bottom: reverse view); (D, E) Conidiophores; (F, G) Conidia (scale bar: 10 μm).
Colony characteristics: On MEA, mycelium floccose, dense, cottony white, greenish-yellow, weakly zonate, margin effuse, white, with poor sporulation and green conidia produced on the aerial mycelium. Sporulation started 26 d after inoculation, the reverse center was yellowish to yellowish brown, and the subcentral margin was yellowish white to colorless. On OA, mycelium floccose, cottony white to green, margin floccose, white to green. Sporulation starts 12 d after inoculation, reverse dark gray, subcentral to margin greenish gray to yellowish brown. On PDA, mycelium floccose, dense, cottony white, margin f loccose, white to pale green. Sporulation started 20 d after inoculation, reverse dark gray, subcentral to margin white.
Conidiophores arising from hyphae, erect, smooth-walled; Phialides solitary borne directly on the hyphae or on conidiophores, smooth-walled, cylindrical, (6–)7–11(–13) × 3–4 μm; Conidia aseptate, smoothwalled, cylindrical, ellipsoidal, usually slightly curved or allantoid, (7–)8–15(–18) × 3–4 μm.
Strains examined: Myeongnye-ri, Hanam-eup, Miryang-si, Gyeongsangnam-do, Republic of Korea, isolated from a cicada cadaver (Cryptotympana atrata), strains CSC23C0001 and CSC23C0002.
Note: Phylogenetically and morphologically, M. viridulum is closely related to M. megapomponiae but differs in conidial size. The conidia of M. viridulum are longer (8–15 × 3–5 μm) compared to those of M. megapomponiae (6–11 × 3–5 μm). On all tested media at 25℃, the Korean strain of M. viridulum exhibited faster growth than the type strain CCRC 32589 (MEA after 14 d: 15–23 mm) [11] and the authentic strain BCC36261 (PDA after 14 d: 15 mm, OA after 14 d: 18 mm) [1].
The authors declare that they have no conflicts of interest.
This research was supported by Changwon National University in 2023–2024 and a grant from the Korea Basic Science Institute (National Research Facilities and Equipment Center) funded by the Ministry of Education (2023R1A6C101B022).
1. Mongkolsamrit S, Khonsanit A, Thanakitpipattana D, Tasanathai K, Noisripoom W, Lamlertthon S, Himaman W, Houbraken J, Samson RA, Luangsa-ard J. Revisiting Metarhizium and the description of new species from Thailand. Stud Mycol 2020;95:171-251. [DOI]
2. Brunner-Mendoza C, Reyes-Montes MdR, Moonjely S, Bidochka MJ, Toriello C. A review on the genus Metarhizium as an entomopathogenic microbial biocontrol agent with emphasis on its use and utility in Mexico. Biocontrol Sci Technol 2019;29:83-102. [DOI]
3. Hu X, Xiao G, Zheng P, Shang Y, Su Y, Zhang X, Liu X, Zhan S, Leger RJS, Wang C. Trajectory and genomic determinants of fungal-pathogen speciation and host adaptation. Proc Natl Acad Sci U S A 2014;111:16796-801. [DOI]
4. National Institute of Biological Resources (NIBR). National species list of Korea [Internet]. Incheon: NIBR; 2024 [cited 2024 Dec 18]. Available from: https://species.nibr.go.kr
5. Rehner SA, Samuels GJ. Taxonomy and phylogeny of Gliocladium analysed from nuclear large subunit ribosomal DNA sequences. Mycol Res 1994;98:625-34. [DOI]
6. Vilgalys R, Hester M. Rapid genetic identification and mapping of enzymatically amplified ribosomal DNA from several Cryptococcus species. J Bacteriol 1990;172:4238-46. [DOI]
7. Rehner SA, Buckley E. A Beauveria phylogeny inferred from nuclear ITS and EF1-α sequences: evidence for cryptic diversification and links to Cordyceps teleomorphs. Mycologia 2005;97:84-98. [DOI]
8. Htet ZH, Hyde KD, Alotibi FO, Chethana TKW, Mapook A. Multigene phylogeny, taxonomy, and potential biological properties of Pseudoroussoella and Neoroussoella species (Roussoellaceae, Dothideomycetes) from Asteraceae weeds in northern Thailand. MycoKeys 2024;111:129-46. [DOI]
9. Tamura K, Peterson D, Peterson N, Stecher G, Nei M, Kumar S. MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol Biol Evol 2011;28:2731-9. [DOI]
10. Katoh K, Standley DM. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol Evol 2013;30:772-80. [DOI]
11. Tzean SS, Hsieh LS, Chen JL, Wu WJ. Nomuraea viridulus, a new entomogenous fungus from Taiwan. Mycologia 1992;84:781-6. [DOI]
12. Stamatakis A. RAxML-VI-HPC: maximum likelihood-based phylogenetic analyses with thousands of taxa and mixed models. Bioinformatics. 2006;22:2688-90. [DOI]