You-Jin Kim1, Seong-Keun Lim1, Song-Woon Nam1, Chang-Gi Back2, Leonid N. Ten3, Seung-Yeol Lee1,3, and Hee-Young Jung1,3*
1Department of Plant Medicine, Kyungpook National University, Daegu 41566, Korea
2Department of Environmental Horticulture and Landscape Architecture, Environmental Horticulture, Dankook University, Cheonan 31116, Korea
3Institute of Plant Medicine, Kyungpook National University, Daegu 41566, Korea
*Correspondence to heeyoung@knu.ac.kr
Korean Journal of Mycology (Kor J Mycol) 2025 March, Volume 53, Issue 1, pages 21-28.
https://doi.org/10.4489/kjm.2025.53.1.3
Received on January 15, 2025, Revised on March 15, 2025, Accepted on March 17, 2025, Published on Mar 31, 2025.
Copyright © The Korean Society of Mycology.
This is an Open Access article which is freely available under the Creative Commons Attribution-NonCommercial 4.0 International License (CC BY-NC) (https://creativecommons.org/licenses/by-nc/4.0/).
The fungal strain designated as KNUF-21-033 was isolated from a soil sample collected in Gunwi-gun, Daegu, Korea. It resembled Varicosporellopsis shangrilaensis in morphological characteristics, including colony color and the shapes of macroconidia and microconidia. The colonies formed white and cottony mycelium on potato dextrose agar (PDA). The conidiophores were erect, septate, and either unbranched or branched at the base, measuring 21.5–53.5 × 2.5–5.1 µm. The macroconidia and microconidia measured 13.722.6 × 3.8–6.9 µm and 4.0–8.8 × 3.5–7.1 µm, respectively. The obtained sequences of the internal transcribed spacer (ITS) regions and the 28S large subunit (LSU) of the ribosomal RNA, β-tubulin (TUB), and actin (ACT) genes of isolate KNUF-21-033 exhibited high similarity to strains CGMCC 3.21000 and KLF 01 of V. shangrilaensis. The topology of the maximum likelihood phylogenetic tree constructed using the concatenated ITS, LSU, TUB, and ACT sequences confirmed the affiliation of KNUF-21-033 with V. shangrilaensis. Morphological observations and phylogenetic analyses indicated that KNUF-21-033 was indeed a V. shangrilaensis strain. To the best of our knowledge, this is the first report of this fungal species in Korea.
Multilocus sequence analysis, Nectriaceae, Soil-inhabiting fungi, Unreported species, Varicosporellopsis shangrilaensis
The family Nectriaceae within the phylum Ascomycota occupies a broad range of ecological niches. Members of this family are predominantly soil-borne saprobes or plant pathogens, whereas some species parasitize other fungi or insects. Additionally, several species have been reported to be significant human pathogens, some of which produce mycotoxins [1]. Since 1865, at least 69 genera have been described in the family Nectriaceae, and advancements in molecular biology have continued to expand the family to include new genera and species [2]. The representative genera within this family include Xenoacremonium, Paracremonium, and Varicosporellopsis. The genus Varicosporellopsis was first established by Lechat and Fournier (2016) with the description of V. aquatilis isolated from freshwater habitats in southern France [3]. Subsequently, V. americana, discovered in the United States, was described by Crous et al. [4]. Another species, V. shangrilaensis was reported in China and isolated from the rhizosphere soil of Astragalus polycladus [5]. These examples highlight that species of Varicosporellopsis have been isolated from diverse environments, including freshwater, waterlogged wood, and rhizosphere soils, classifying them as environmental microorganisms [3–6]. To date, this genus comprises three described species. Morphologically, the type species of Varicosporellopsis is characterized by macronematous, mononematous, unbranched, flexuous conidiophores with smooth, hyaline walls. Conidiogenous cells are monophialidic, featuring a slightly flared collarette, and produce narrowly ellipsoidal to subcylindrical, smooth, hyaline conidia. The establishment of the three above-mentioned members of the genus Varicosporellopsis was performed by applying phylogenetic analysis based on molecular markers such as internal transcribed spacer (ITS) regions and the 28S large subunit (LSU) of the ribosomal RNA, β-tubulin (TUB), and actin (ACT) genes [3–6].
The aim of this study was to explore the diversity of indigenous fungal species in Korea. As part of this effort, a fungal strain, designated KNUF-21-033, was isolated from soil in Korea. This study aimed to identify and characterize previously unreported fungal species belonging to the genus Varicosporellopsis. Morphological and molecular analyses were conducted to identify the fungal strains, and the findings are presented below.
A soil sample was collected from Gunwi-gun, Daegu, Korea (35°59’33.9″N 128°41’12.7″E) and brought to the laboratory. Fungal strains were isolated using serial dilutions. One gram of each soil sample was mixed with 10 mL of sterile distilled water, vortexed, serially diluted, and spread onto potato dextrose agar (PDA; Difco, Detroit, MI, USA) plates. All plates were incubated at 25°C for 7 days. Well-growing colonies were isolated, cultured on fresh PDA plates, and incubated under the same conditions. Several fungal strains were isolated and initially identified using ITS sequences. As a result, isolate KNUF-21033 was preliminarily identified as a fungal species not previously reported in Korea. Further cultural, morphological, and molecular phylogenetic analyses were conducted to identify the isolates. Isolate KNUF21-033 was deposited at the National Institute of Biological Resources (NIBR) as a metabolically inactive culture (NIBRFGC000509258).
For morphological analysis, isolate KNUF-21-033 was incubated at 25°C on PDA for 10 days [5]. After 10 days, the morphological characteristics of the strain, such as color, size, and shape, were recorded. Observations, measurements, and photographs of the colonies, conidia, conidiogenous cells, and conidiophores were taken using a light microscope (BX-50, Olympus, Tokyo, Japan).
Total genomic DNA was extracted using the HiGene Genomic DNA Prep Kit (BIOFACT, Daejeon, Korea) following the manufacturer’s instructions. For molecular analysis, ITS regions and LSU, TUB, and ACT genes were amplified via PCR. PCR amplification was performed using primer pairs ITS1/ITS4 for the ITS regions [7,8], LR0R/LR7 for LSU [9], T1/T22 for TUB [10], and ACT512F/ACT1Rd for ACT [11,12]. The amplification conditions for each gene have been previously described [5].
A phylogenetic tree was constructed using ITS, LSU, TUB, and ACT sequences for strain KNUF21-033, along with closely related strains whose sequences were retrieved from the National Center for Biotechnology Information (NCBI) GenBank database. Basic Local Alignment Search Tool (BLAST) was used for sequence comparison. Twelve taxa were included in the phylogenetic analysis (Table 1). The sequences, including those of KNUF-21-033, were aligned to each molecular marker. The aligned datasets for each strain were concatenated in the following order: ITS regions, LSU, TUB, and ACT genes. The Kimura model was used to estimate evolutionary distances and distance matrices were generated based on these calculations [13]. A phylogenetic tree was constructed using the maximum likelihood (ML) method in MEGA 11.0 software and bootstrapped with 1,000 replicates to ensure statistical robustness [14,15].
Table 1. List of species used in phylogenetic analysis along with their GenBank accession numbers
| Species | Strain | GenBank accession numbers | |||
|---|---|---|---|---|---|
| ITS | LSU | TUB | ACT | ||
| Corallomycetella elegans | CBS 275.60 | KM231828 | KM231710 | KM232100 | KM231237 |
| Microcera coccophila | MAFF 241482 | KC291752 | KC291787 | KC291936 | – |
| Paracremonium contagium | CBS 110348 | NR_154313 | NG_058130 | KM232103 | KM231240 |
| Paracremonium inflatum | CBS 485.77 | NR_154312 | NG_058129 | KM232101 | KM231238 |
| Varicosporellopsis americana | CBS 148257T | NR_175234 | NG_081341 | OK651211 | OK651131 |
| Varicosporellopsis aquatilis | CBS 143509 | MH107922 | MH107968 | MH108052 | MH107987 |
| Varicosporellopsis shangrilaensis | CGMCC 3.21000T | OM956089 | OP223500 | OQ658529 | OQ658531 |
| Varicosporellopsis shangrilaensis | KNUF-21-033 | LC858579 | LC858580 | LC858581 | LC858582 |
| Varicosporellopsis shangrilaensis | KLF 01 | OP223502 | OP223501 | OQ658530 | OQ658532 |
| Xenoacremonium falcatus | CBS 400.85 | KM231832 | HQ232025 | KM232104 | – |
| Xenoacremonium recifei | CBS 137.35T | KM231833 | HQ232106 | KM232105 | KM231241 |
| Atractium crassum | CBS 180.31T | KM231790 | MH866623 | KM232049 | HQ897859 |
ITS: internal transcribed spacer regions; LSU: the 28S large subunit of the ribosomal RNA gene; TUB: β-tubulin gene; ACT: actin gene.
T Type strain. The strain isolated in this study is indicated in boldface.
After 10 days of incubation, the colony diameter of strain KNUF-21-033 reached 26.0–28.0 mm and exhibited a white, cottony surface (Fig. 1A and B). After more than two weeks of incubation, the colonies gradually turned pale yellow. The conidiophores of the isolate measured 21.5–53.5 × 2.5–5.1 µm, were straight, segmented, and either unbranched or branched at the base. Its conidiogenous cells measured 2.9–7.7 × 3.1–5.9 µm and featured a diminutive collarette, bearing small conidia at the apex. Strain KNUF-21-033 produced two types of conidia: macroconidia and microconidia. The macroconidia were kidney-shaped or boat-shaped, linear or curved, guttulate, and measured 13.7–22.6 × 3.8–6.9 µm. The microconidia were spherical or droplet-shaped, guttulate, and measured 4.0–8.8 × 3.5–7.1 µm (Fig. 1C–F). The cultural and morphological characteristics of strain KNUF-21-033 closely resembled those of V. shangrilaensis [5] (Table 2). However, as shown in Table 2, strain KNUF-21-033 can be distinguished from its close relative V. americana by several morphological traits. The conidiophores of V. americana are larger (40.0–70.0 × 3.0–4.0 µm), solitary, and branched at the base. Additionally, the conidiogenous cells of V. americana are significantly larger (30.0–50.0 × 3.0–4.0 µm), subcylindrical, and feature a flared collarette that produces slimy, hyaline, ellipsoid conidia, with a subobtuse apex and a tapered base. These conidiogenous cells and conidia differed markedly from those of strain KNUF-21-033 in terms of size and shape.
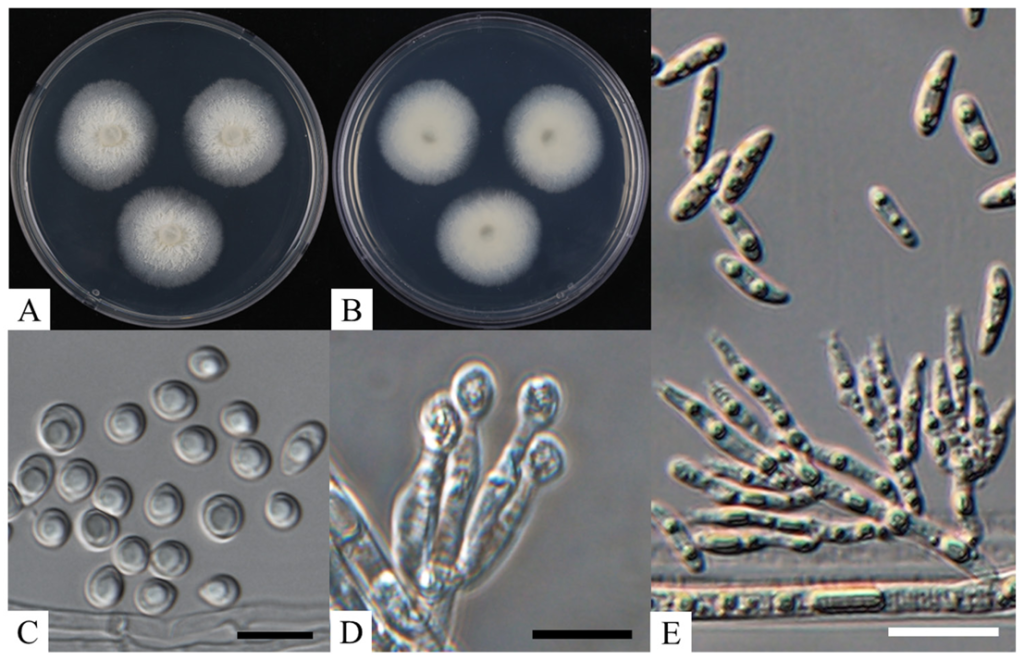
Fig. 1. Culture and morphological characteristics of KNUF-21-033. A: surface of the colony on potato dextrose agar (PDA); B: reverse of the colony on PDA; C: microconidia; D: conidiogenous cell and conidiophore; E: macroconidia, conidiogenous cell, and conidiophore (Scale bar C, D = 10 µm, E = 20 µm).
Table 2. Comparison of the morphological characteristics of strain KNUF-21-033 with those of previously reported Varicosporellopsis species
| Characteristics | Varicosporellopsis shangrilaensisa KNUF-21-033 | V. shangrilaensisb CGMCC 3.21000 | V. americanac CBS 143509 | |
|---|---|---|---|---|
| Colony | Color | White, gradually turning to pale yellow | Initially white, gradually turning to pale yellow | Saffron on both the surface and reverse |
| Size (diam) | 26.0–28.0 mm in 10 days at 25℃ | 30 mm in 10 days at 26℃ | 30 mm in 14 days at 25℃ | |
| Shape | Cottony | Cottony | Fat, smooth, and lobate margin | |
| Conidiophores | Size (µm) | 21.5–53.5 × 2.5–5.1 | N/A | 40.0–70.0 × 3.0–4.0 |
| Shape | Straight, segmented, either unbranched or branched at the base | Macronematous, erect, septate, unbranched or branched at the base | Solitary, erect, branched at base, 0–2-septate cells | |
| Conidiogenous cell | Size (µm) | 2.9–7.7 × 3.1–5.9 | 10.0–20.0 × 3.0–5.0 | 30.0–50.0 × 3.0–4.0 |
| Shape | Phialidic with a diminutive collarette, bearing small conidia at the apex | Phialidic with a minute collarette | Subcylindrical with slight apical taper, hyaline, smooth, apex phialidic with flared collarette, giving rise to clusters of slimy conidia | |
| Conidia | Size (µm) | Macroconidia: 13.7–22.6 × 3.8–6.9 Microconidia: 4.0–8.8 × 3.5–7.1 | Macroconidia: 8.0–20.0 × 2.5–8.5 Microconidia: 3.5–6.5 × 3–6 | (10 –)12–16(–24) × (3–)4(–4.5) |
| Shape | Macroconidia: kidney-shaped or boat-shaped, linear or curved, guttulate Microconidia: round or droplet-shaped, guttulate | Macroconidia: reniform or cymbiform, straight or slightly curved, guttulate Microconidia: spherical or t eardrop-shaped, guttulate | Solitary, hyaline, smooth, guttulate, ellipsoid, aseptate, straight to curved, apex subobtuse, base tapered to a truncate hilum |
PDA: potato dextrose agar; diam: diameter.
a Fungal strain used in this study; b Source of the description [5]; c Source of the description [4].
To identify strain KNUF-21-033 at the species level, the nucleotide sequences of the ITS, LSU, TUB, and ACT loci were obtained (572, 1328, 1332, and 1191 bp, respectively) and compared with those of other strains in the GenBank database using BLAST. The ITS sequence of the isolate exhibited 99.8% similarity with those of two V. shangrilaensis strains, CGMCC 3.21000 (OM956089) and KLF 01 (OP223502); 96.0% similarity with V. americana CBS 148257 (NR_175234); and a significantly lower similarity of 92.9% with Paracremonium inflatum CBS 485.77 (NR_154312). The LSU sequence showed 99.9% similarity with the two V. shangrilaensis strains, CGMCC 3.21000 (OP223500) and KLF 01 (OP223501), and 97.7% similarity with several strains of Fusicolla quarantenae, including GZUIFR 21.906 (OL897056), GZUIFR 21.907 (OL897057), and CGMCC 3.20777 (OL897054). Unfortunately, BLAST searches for the TUB and ACT gene sequences of strain KNUF-21-033 did not return results for the aforementioned Varicosporellopsis strains. Therefore, similarity values for the TUB and ACT sequences were calculated via pairwise comparisons. The TUB sequence of strain KNUF-21-033 exhibited 99.6% similarity with V. shangrilaensis CGMCC 3.21000 (OQ658529) and KLF 01 (OQ658530) and only 92.3% similarity with V. americana CBS 148257 (OK651211). The ACT sequence showed 96.2% similarity with V. shangrilaensis CGMCC 3.21000 (OQ658531), 96.9% similarity with KLF 01 (OQ658532), and 95.5% similarity with V. americana CBS 148257 (OQ651131). Based on these comparisons, it was evident that the use of a single molecular marker is insufficient for accurate species-level identification. Therefore, a multilocus sequence analysis (MLSA) was performed. For the establishment of the genus Varicosporellopsis, ITS and LSU sequences were initially used [6], while more recently, ITS, LSU, TUB, and ACT sequences were utilized to establish V. shangrilaensis [5]. Following the latter approach, MLSA was performed using concatenated sequences of the ITS, LSU, TUB, and ACT loci. The combined sequences were used to construct an ML phylogenetic tree (Fig. 2). The resulting topology clearly indicated that three strains, KNUF-21-033, CGMCC 3.21000, and KLF 01, belonged to V. shangrilaensis. The results of both morphological and phylogenetic analyses identified strain KNUF-21-033 as V. shangrilaensis. To the best of our knowledge, this is the first report on this fungal species in Korea.
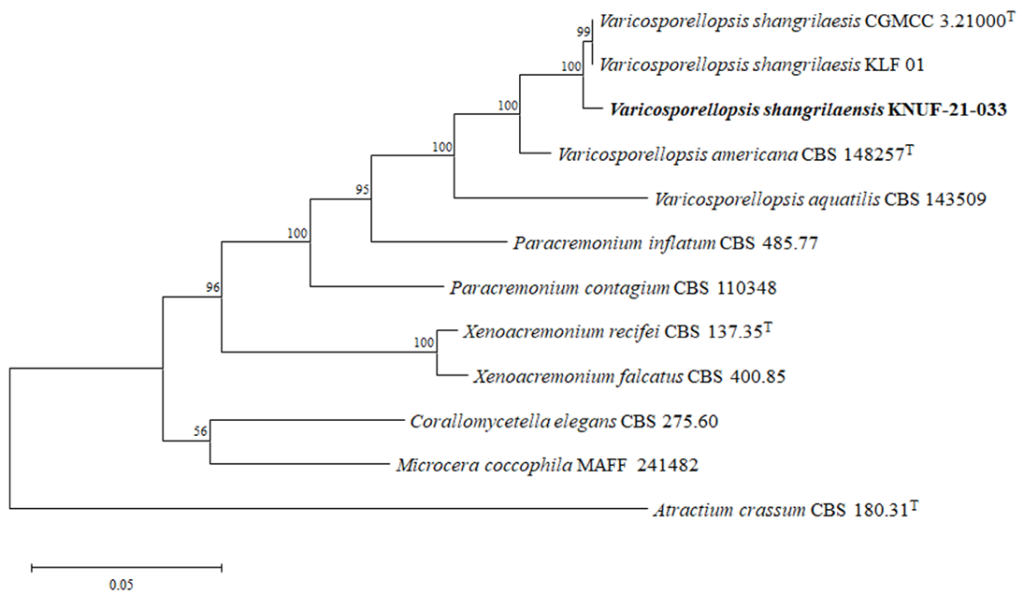
Fig. 2. Maximum likelihood phylogenetic tree based on a combined dataset of particle sequences of internal transcribed spacer (ITS) regions and the 28S large subunit (LSU), β-tubulin (TUB) and actin (ACT) genes. Atractium crassum CBS 180.31T was used as an out-group. The strain isolated in this study is in bold, and the bootstrap values are based on 1,000 replications. Bar, 0.05 substitutions per nucleotide position.
Members of the genus Varicosporellopsis have been isolated from submerged wood in freshwater environments and rhizosphere soil and are considered environmental microorganisms [3–6]. Molecular phylogenetic analyses showed that Varicosporellopsis is closely related to the genera Paracremonium, Xenoacremonium, and Corallomycetella [2], with Paracremonium being the closest relative. Notable species of Paracremonium include P. inflatum and P. contagium, which were isolated from granulomatous and subcutaneous lesions in humans in India and Canada, respectively [1]. Additionally, P. pembeum was recovered from an ambrosia beetle (Euwallacea sp.), and P. lepidopterorum was isolated from a lepidopteran pupa [16,17]. Although Varicosporellopsis species are generally regarded as environmental microorganisms, the discovery of a pathogenic Paracremonium species isolated from human lesions underscores their potential medical significance of Varicosporellopsis. These findings highlight the need for detailed studies on the distribution, biological activities, and potential pathogenicity of Varicosporellopsis spp. Moreover, reports on Paracremonium species forming symbiotic relationships with insects suggest that Varicosporellopsis may exhibit similar associations. Despite its recent establishment as a genus, Varicosporellopsis currently includes only three described species [3–6], and no substantial follow-up studies have been conducted on them. This limited knowledge emphasizes the importance of exploring new members of this genus and studying the ecological, etiological, and biological roles of the known species. Beyond the documented sources of its isolation and its cultural, morphological, and phylogenetic traits, little information is available regarding this genus. Further investigation is essential to achieve a more comprehensive understanding of Varicosporellopsis and its species.
This study is the first report of V. shangrilaensis infection in Korea. The isolated strain, KNUF-21033, identified as a member of V. shangrilaensis, provides a valuable resource for future research on this species in Korea. Its isolation offers an opportunity to deepen our insight into the ecological, etiological, and biological roles of V. shangrilaensis, particularly under the unique environmental conditions of Korea. This study also contributes to enhancing the understanding of the fungal diversity indigenous to Korea, underscoring the need for continued exploration of the country’s fungal biodiversity.
The authors declare that they have no potential conflicts of interest.
This study was supported by the National Institute of Biological Resources funded by the Ministry of Environment of the Republic of Korea (NIBR202102107).
1. Lombard L, van der Merwe NA, Groenewald JZ, Crous PW. Generic concepts in Nectriaceae. Stud Mycol 2015;80:189-245. [DOI]
2. Li X, Han SL, Zhang YY, Cai L, Zhao P. Heteroverticillium phytelephatis gen. et sp. nov. intercepted from nuts of Phytelephas macrocarpa, with an updated phylogenetic assessment of Nectriaceae. Mycology 2023;14:155-74. [DOI]
3. Lechat C, Fournier J. Varicosporellopsis, a new aquatic genus from southern France. Ascomycete.org 2016;8:96-100.
4. Crous PW, Osieck ER, Jurjević Ž, Boers J, van Iperen AL, Starink-Willemse M, Dima B, Balashov S, Bulgakov TS, Johnston PR, et al. Fungal planet description sheets: 1284-382. Persoonia 2021;47:178-374. [DOI]
5. Dou ZR, Zhang F, Zhou GJ, Gong GF, Ding JJ, Fan XH, Jiang B, Wang K. Varicosporellopsis shangrilaensis sp. nov. (Nectriaceae, Hypocreales), a new terricolous species isolated from the rhizosphere soil of Astragalus polycladus in Northwestern Yunnan, China. Phytotaxa 2023;600:230-38. [DOI]
6. Lechat C, Fournier J. Varicosporella, a new aquatic genus in the Nectriaceae from France. Ascomycete.org 2015;7:1-8.
7. White TJ, Bruns TD, Lee SB, Taylor JW. Amplification, and direct sequencing of fungal ribosomal RNA genes for phylogenetics. In: Innis MA, Gelfand DH, Sninsky JJ, White TJ, editors. PCR protocols: a guide to methods and applications. New York: Academic Press; 1990. p. 315-22. [DOI]
8. Gardes M, Bruns TD. ITS primers with enhanced specificity for basidiomycetes-application to the identification of mycorrhizae and rusts. Mol Ecol 1993;2:113-8. [DOI]
9. Bunyard BA, Nicholson MS, Royse DJ. A systematic assessment of Morchella using RFLP analysis of the 28S ribosomal RNA gene. Mycologia 1994;86:762-72. [DOI]
10. Bills GF, Platas G, Overy DP, Collado J, Fillola A, Jiménez MR. Martín J, del Val AG, Vicente F, Tormo JR, et al. Discovery of the parnafungins, antifungal metabolites that inhibit mRNA polyadenylation, from the Fusarium larvarum complex and other Hypocrealean fungi. Mycologia 2009;101:449-72. [DOI]
11. Cabral A, Rego C, Nascimento T, Oliveira H, Groenewald JZ, Crous PW. Multi-gene analysis and morphology reveal novel Ilyonectria species associated with black foot disease of grapevines. Fungal Biol 2009;116:62-80. [DOI]
12. Groenewald JZ, Nakashima C, Nishikawa J, Shin HD, Park JH, Jama AN, Groenewald M, Braun U, Crous PW. Species concepts in Cercospora: spotting the weeds among the roses. Stud Mycol 2013;75:115-70. [DOI]
13. Kimura M. A simple method for estimating evolutionary rates of base substitutions through comparative studies of nucleotide sequences. J Mol Evol 1980;16:111-20. [DOI]
14. Fitch WM. Toward defining the course of evolution: minimum change for a specific tree topology. Syst Zool 1971;20:406-16. [DOI]
15. Tamura K, Stecher G, Kumar S. MEGA11: Molecular evolutionary genetics analysis version 11. Mol Biol Evol 2021;38:3022-7. [DOI]
16. Lynch SC, Twizeyimana M, Mayorquin JS, Wang DH, Na F, Kayim M, Kasson MT, Thu PQ, Bateman C, Rugman-Jones P, et al. Identification, pathogenicity and abundance of Paracremonium pembeum sp. nov. and Graphium euwallaceae sp. nov.-two newly discovered mycangial associates of the polyphagous shot hole borer (Euwallacea sp.) in California. Mycologia 2016;108:313-29 [DOI]
17. Ming DQ, Luo LY, He XX, Wang MS, Fang WX, Chen SF, Chen WH, Han YF, Liang ZQ. Paracremonium lepidopterorum, a new insect-associated fungus. Phytotaxa 2021;524:85-91. [DOI]