Muhammad Saeed1*, Sakshi Sharma1, Stefan Seegmüller2, and Matthias Hahn1
1RPTU Kaiserslautern-Landau, Department of Biology, 67663 Kaiserslautern, Germany
2Research Institute for Forest Ecology and Forestry Rheinland-Pfalz, 67705 Trippstadt, Germany
*Correspondence to saeed@rptu.de
Korean Journal of Mycology (Kor J Mycol) 2025 March, Volume 53, Issue 1, pages 29-35.
https://doi.org/10.4489/kjm.2025.53.1.4
Received on February 07, 2025, Revised on March 14, 2025, Accepted on March 17, 2025, Published on Mar 31, 2025.
Copyright © The Korean Society of Mycology.
This is an Open Access article which is freely available under the Creative Commons Attribution-NonCommercial 4.0 International License (CC BY-NC) (https://creativecommons.org/licenses/by-nc/4.0/).
Oak powdery mildew (OPM) is a major disease of oak trees native in central Europe. In contrast, red oak (Quercus rubra) is generally considered immune to OPM. In 2024, five instances of powdery mildew infection were observed on Q. rubra in Kaiserslautern, Germany. Molecular analyses identified the pathogens as either Erysiphe alphitoides or E. quercicola. Controlled inoculations of Q. rubra leaves with E. alphitoides inoculum obtained from Q. rubra showed very limited pathogen development. Few germinated conidia formed secondary hyphae with haustoria, and no mycelium was observed, suggesting nonhost resistance to OPM. While Q. rubra appears resistant to OPM, these sporadic reports suggest continuous monitoring to better understand this potential disease threat for an important tree species.
Erysiphe alphitoides, E. quercicola, Nonhost resistance, Northern red oak, Oak powdery mildew
Oak powdery mildew (OPM), caused by Erysiphe spp., is the most significant leaf disease on native oaks in Europe and represents one of the primary obstacles to the successful rejuvenation of oak trees in forests. Three cryptic OPM species are responsible for the disease: Erysiphe alphitoides, E. quercicola and E. hypophylla. Among these, E. alphitoides is the most widespread across European countries, while E. quercicola has a limited distribution and E. hypophylla is quite rare [1]. Quercus rubra L., commonly known as northern red oak, is native to North America and is cultivated as an ornamental tree in temperate regions in Europe. Covering about 55,000 hectares (0.5%) of the total forest area, Q. rubra is the most prevalent non-native broad-leaved tree species in Germany [2]. The two native species in Germany, Q. robur (pedunculate oak) and Q. petraea (sessile oak), are both susceptible, but Q. rubra has been found to be generally immune to OPM [3,4]. However, E. quercicola was reported for the first time as causing powdery mildew on several Q. rubra trees in a public garden in Seoul, Korea [5].
In February and April 2024, two Q. rubra seedlings growing in our phytochamber exhibited OPM symptoms (Table 1: samples 1 and 2; Fig. 1A: sample 1). The fungal mycelium was growing on the upper leaf surface, characteristic of majority of powdery mildew diseases [6]. In July 2024, one tree (Table 1: sample 3) growing on the campus of the RPTU in Kaiserslautern was found that showed typical OPM symptoms with profuse white mycelium on the upper surfaces of the newly grown leaves, whereas the older leaves showed no or only weak symptoms (Fig. 1B). These infected leaves (samples 1–3) were stained with trypan blue (Acros Organics BV – Geel, Belgium). For staining, the infected Q. rubra leaves were placed overnight in a clearing solution (acetic acid, ethanol, glycerol, 1:3:1). The solution was replaced with staining solution (0.05% trypan blue in 1:1:1 lactic acid, glycerol, water), and shaken at 50 rpm overnight. After staining, the leaf samples were rinsed thrice with water and mounted on glass slides. Microscopy of trypan blue-stained leaves (samples 1–3) revealed fungal mycelium only on the upper leaf surface (Fig. 1C).
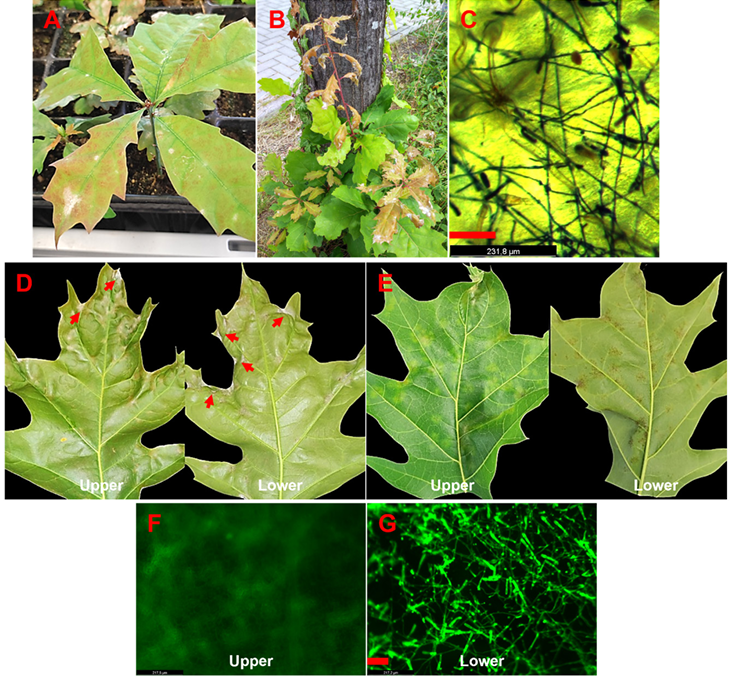
Fig. 1. Quercus rubra seedling from a phytochamber, displaying symptoms of powdery mildew (sample 1, A). Q. rubra tree growing on the RPTU campus, showing symptoms of powdery mildew (sample 3, B). Trypan blue staining revealing a mycelium network on the upper leaf surface of Q. rubra (sample 3, C). Q. rubra leaves from a plant growing on the RPTU campus with powdery mildew-like symptoms (highlighted by arrows, sample 4, D), and leaf discoloration and mottling on upper side and corresponding brown spotting on the lower side (sample 4, E). Fluorescent staining with wheat germ agglutinin Oregon GreenTM 488 conjugate reveals sporulating mycelium on the Q. rubra leaf shown in panel D (F and G). Bars represent 100 µm.
In September 2024, several large and mature leaves from two other Q. rubra plants (samples 4 and 5) displayed a thin layer of powdery mildew-like mycelium on both leaf surfaces (Fig. 1D, type 1 symptoms). Other leaves from the same plants showed light green mottling on the upper surface, suggesting early mildew infection or chlorosis, while the lower surface displayed brown spots without visible mycelium (Fig. 1E, type 2 symptoms). Both types of infected leaves were stained with trypan blue and observed under bright field microscopy. Type 1 symptomatic leaf samples showed fungal mycelium on both the upper and lower leaf surfaces, whereas type 2 symptomatic leaf samples revealed weakly developed fungal mycelium exclusively on the lower leaf surface (not shown). To improve the visualization of type 2 symptomatic leaves (samples 4 and 5), they were additionally stained with wheat germ agglutinin (WGA) Oregon GreenTM 488 conjugate (Thermo Fisher Scientific, Langenselbold, Germany). For staining, infected leaves were placed in 5% KOH in a shaker at 100 rpm at 37°C overnight. The KOH solution was removed, the leaves were washed with 1× PBS buffer, transferred to WGA solution (10 µg/ml in 1× PBS buffer), vacuum infiltrated for 15 minutes, and incubated in the dark at 4°C on a shaker at 50 rpm overnight. After staining, the solution was discarded, and leaf samples were rinsed three times with 1× PBS. Samples were mounted on glass slides and observed with a GFP filter using a Leica M205 FCA fluorescence stereo microscope (Leica Microsystems, Wetzlar, Germany). Microscopy of type 2 symptomatic leaf samples confirmed fungal mycelium to be present exclusively on the lower leaf surface (Fig. 1F and 1G).
To identify the powdery mildew species responsible for Q. rubra infection, total DNA was extracted from leaf samples using the miniprep CTAB-based method described by Untergasser [7], and the internal transcribed spacer (ITS) and large subunit (LSU) regions were amplified using conserved powdery mildewspecific primers AITS/TW14 [8]. The resulting amplicons were sequenced using primers ITS4 and ITS5 [9] for the ITS region, as well as PM28R [8] and LROR [10] for the LSU region. The sequences obtained were compared against reference databases. Sequencing confirmed the presence of E. alphitoides on three plants and E. quercicola on one plant, and both E. alphitoides and E. quercicola on the same leaf of another plant (Table 1). The ITS+LSU sequences of all four E. alphitoides samples were identical to each other, and the ITS+LSU sequences of the two E. quercicola samples were also identical. Additional ITS+LSU sequences from Erysiphe species on Quercus spp. and other closely related powdery mildew species were retrieved from NCBI (National Center for Biotechnology Information), aligned using MUSCLE (Multiple Sequence Comparison by Log- Expectation) in MEGA11 [11], and a phylogenetic tree of these sequences was constructed by using the neighbour-joining method (Fig. 2). This analysis confirmed the close relationship of the E. alphitoides and E. quercicola isolates from Germany with other known Erysiphe spp., supporting the identification of these species on Q. rubra. The ITS+LSU sequences from each species were deposited in GenBank under accession numbers PQ094592 (sample 3, E. alphitoides) and PQ094239 (sample 1 E. quercicola).
Table 1. Powdery mildew-infected samples from Quercus rubra
| Samples | Origin (GPS) | Species | Collection date |
|---|---|---|---|
| 1 | Phytochamber, Trippstadt (next to Kaiserslautern), (49.3541, 7.7671) | E. quercicola | Feb, 2024 |
| 2 | E. alphitoides | Apr, 2024 | |
| 3 | RPTU Campus, Kaiserslautern, (49.4250, 7.7548) | E. alphitoides | July, 2024 |
| 4 | E. alphitoides | Sep, 2024 | |
| 5 | E. alphitoides, E. quercicola | Sep, 2024 |
GPS: Global Positioning System.
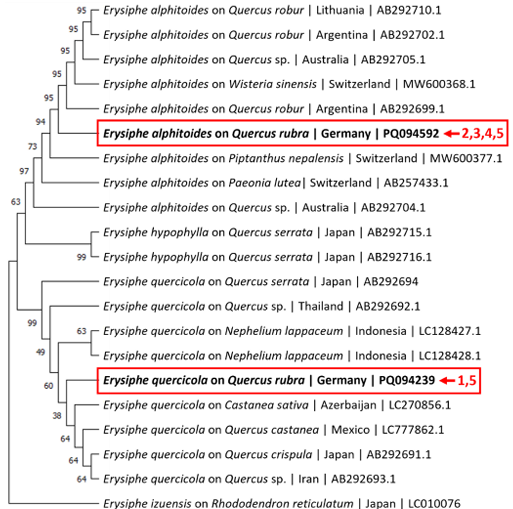
Fig. 2. Neighbour-joining tree depicting selected sequences from the internal transcribed spacer (ITS) and large subunit (LSU) regions. The sequences marked with arrows were obtained for the Quercus rubra plants in this study. Sample numbers are corresponding to Table 1. Erysiphe izuensis was selected as an outgroup taxon.
The low number of conidia obtained from the E. quercicola infection precluded their use for inoculations; however, sufficient conidia from two separate E. alphitoides-infected Q. rubra plants were collected. Young Q. rubra leaves (n = 18; sample 1: n = 2, sample 3: n = 16), and Q. petraea (n = 6) control leaves were inoculated with two conidia suspensions of E. alphitoides, one obtained from Q. rubra sample 1 and the other from strongly infected sample 3. With both suspensions, Q. petraea oak leaves developed visible mycelium by 3 days post-inoculation (dpi), which progressively expanded (Fig. 3A). In contrast, no visible white fungal mycelium was detected on Q. rubra leaves even by 12 dpi (Fig. 3B). When the inoculum from sample 3 was used, out of 16 Q. rubra leaves, two showed rudimentary mycelium-like appearance at 12 dpi (Fig. 3C). In both infection experiments, the identity of E. alphitoides in the inoculum, as well as in the mycelium developed on sessile oak and the rudimentary mycelium on Q. rubra, was confirmed by sequencing. These observations showed that controlled inoculation with E. alphitoides conidia obtained from infected Q. rubra failed to induce bona fide OPM symptoms in Q. rubra upon reinfection, confirming the inherent resistance of Q. rubra to OPM.
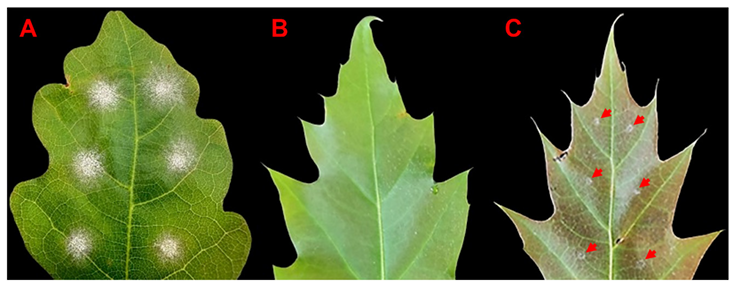
Fig. 3. Results of controlled inoculations of Quercus petraea (A) and Q. rubra (B, C) leaves with Erysiphe alphitoides conidia isolated from a sporulating lesion on Q. petraea (sample 3). Whereas strong powdery mildew infection was observed in susceptible Q. petraea (A), most of the Q. rubra leaves showed no symptoms (B), except for two Q. rubra leaves displaying rudimentary mycelium appearance, highlighted by arrows (C). Pictures were taken at 12 dpi.
Microscopic examination of Q. rubra leaves inoculated with E. alphitoides conidia collected from sample 3 revealed that at 2 dpi most conidia had geminated but did not progress beyond appressoria formation stage. However, approximately 9% of the germinated conidia (n = 200) had formed elongating secondary hyphae (ESH) (Fig. 4A). The mean length of ESH (n = 27) was 105 ± 34 µm. In contrast, an extensive network of ESH was observed on Q. petraea leaves at 2 dpi (Fig. 4B). By 5 dpi, some growth of ESH was observed in Q. rubra leaves, with a mean length of ESH units (n = 10) measuring 366 ± 64, while extensive conidiophore production was noted only in Q. petraea leaves. At 12 dpi, Q. rubra leaves with rudimentary mycelium exhibited a sparse ESH network in all 12 observed spots, with no conidiophore formation (Fig. 4C). In contrast, Q. rubra leaves without visible mycelium displayed a less developed, sparse ESH network in only 3 of 16 observed spots. These findings highlight the limited development of E. alphitoides on Q. rubra, supporting its resistance to the disease.
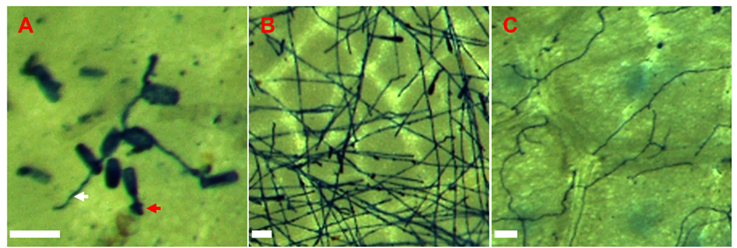
Fig. 4. Trypan blue staining of a Quercus rubra leaf (pictured in Fig. 3C), showing germinated conidia forming appressoria (red arrow) and rudimentary hyphae (white arrow) only (A), compared to the extensive hyphal network in susceptible control Q. petraea at 2 dpi (B). Weak development of hyphae on Q. rubra at 12 dpi (C). Bars represent 50 µm.
Although it was reported that E. quercicola caused powdery mildew on Q. rubra trees, the study did not include controlled inoculations on Q. rubra [5]. Consequently, Koch’s postulates, which are essential for confirming E. quercicola as the causal organism on Q. rubra, remained unfulfilled.
Over a three-year period (September 2021 to September 2024), we collected 53 samples of powdery mildew-infected oak leaves in the forest region close to Kaiserslautern. Total DNA was extracted, and the entire ITS region was amplified using the Erysiphales-specific primers PMITS1/PMITS2 [12]. Sequencing of 20 samples was performed using nested primers ITS4 or ITS5 [9]. Additionally, PCR products from 33 samples amplified with PMITS1/PMITS2 were reamplified using the nested ITS4/ITS5 primers. The resulting products were digested with PvuII, BsrI, or BsmAI to confirm the identity of OPM species, following the protocol described in [13]. We identified E. alphitoides in most cases (49), while E. quercicola was detected in only four cases. In this study a similar prevalence of E. alphitoides and E. quercicola was found on Q. rubra, emphasizing that either species can sporadically infect this host.
Our observations partially align with the concept of nonhost resistance of Q. rubra against E. alphitoides. It remains to be investigated why some Q. rubra trees, such as those in a park in Korea [5] and those in our study in Germany, developed well-established, sporulating colonies of OPM which indicate susceptibility of these Q. rubra plants. Given these isolated instances and the failure of the OPM conidia isolated from Q. rubra leaves to reinfect Q. rubra we suggest considering them as sporadic occurrences for the time being. Nevertheless, it cannot be excluded that our inoculation method which works well with detached leaves of the susceptible Q. petraea is not efficient for Q. rubra. However, these observations warrant ongoing monitoring to better understand this potential threat.
None.
This project has been funded by the Fachagentur Nachwachsende Rohstoffe e.V. (BMEL), ref. no. 2220WK1684 (2021–2024).
1. Desprez-Loustau ML, Massot M, Toïgo M, Fort T, Kaya AGA, Boberg J, Braun U, Capdevielle X, Cech T, Chandelier A, et al. From leaf to continent: the multi-scale distribution of an invasive cryptic pathogen complex on oak. Fungal Ecol 2018;36:39-50. [DOI]
2. BMEL. Dritte Bundeswaldinventur 2012 [Internet]. Berlin: Bundesministerium für Ernährung und Landwirtschaft; 2012 [cited 2017 Nov 8]. Available from https://www.bundeswaldinventur. de/dritte-bundeswaldinventur-2012.
3. Edwards MC, Ayres PG. Cell death and cell wall papillae in the resistance of oak species to powdery mildew disease. New Phytol 1981;89:411-8. [DOI]
4. Marçais B, Desprez-Loustau ML. European oak powdery mildew: impact on trees, effects of environmental factors, and potential effects of climate change. Ann For Sci 2014;71:633-42. [DOI]
5. Choi YJ, Park JH, Choi IY, Abasova L, Choi JH, Shin HD. Erysiphe quercicola causing powdery mildew on Quercus rubra in Korea. Kor J Mycol 2022;50:137-41.
6. Boddy L. Pathogens of autotrophs. In: Watkinson SC, Boddy L, Money NP, editors. The Fungi, 3rd edition. New York: Academic Press; 2016. p. 245-92. [DOI]
7. Untergasser A. 2008. DNA miniprep using CTAB [Internet]. Publisher unknown [cited 2025 Mar 20]. Available from https://www.untergasser.de/lab/protocols/miniprep_dna_ctab_v1_0. pdf.
8. Bradshaw M, Tobin PC. Sequencing herbarium specimens of a common detrimental plant disease (powdery mildew). Phytopathology 2020;110:1248-54. [DOI]
9. White TJ, Bruns TD, Lee SB, Taylor JW. Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics. In: Innis MA, Gelfand DH, Sninsky JJ, White TJ, editors. PCR – protocols and applications: a laboratory manual. New York: Academic Press; 1990. p. 315-22. [DOI]
10. Vilgalys R, Hester M. Rapid genetic identification and mapping of enzymatically amplified ribosomal DNA from several Cryptococcus species. J Bacteriol 1990;172:4238-46. [DOI]
11. Tamura K, Stecher G, Kumar S. MEGA11: Molecular evolutionary genetics analysis version 11. Mol Biol Evol 2021;38:3022-7. [DOI]
12. Cunnington JH, Takamatsu S, Lawrie AC, Pascoe IG. Molecular identification of anamorphic powdery mildews (Erysiphales). Australasian Plant Pathol 2003;32:421-8. [DOI]
13. Feau N, Lauron-Moreau A, Piou D, Marçais B, Dutech C, Desprez-Loustau ML. Niche partitioning of the genetic lineages of the oak powdery mildew complex. Fungal Ecol 2012;5:154-62. [DOI]