Section
Mt. Taebaeksan extends from Gangwon-do Province (Taebaek-si, Yeongwol-gun, and Jeongseon-gun) to Gyeongsangbuk-do Province (Bongwha-gun) in South Korea and has an area of 70,052 km2. Of Korea’s 22 national parks, Mt. Taebaeksan is the most recently designated. Taebaeksan National Park is home to a variety of wildlife, including several endangered species, such as Egretta eulophotes Swinhoe, Pelophylax chosenicus Okada, and Lutra lutra L. The higher plains are covered by alpine vegetation. Although macrofungi surveys have been conducted in other national parks with support from the national government, little research has been conducted on macrofungi in the Taebaeksan National Park. Nevertheless, because the vegetation of the park is diverse, a variety of macrofungi was expected to be distributed throughout. Through research done in the park from 2019 to 2022, 999 specimens of macrofungi were obtained, first identified based on their macroscopic morphology, followed by accurate identification through microstructural observation using a microscope. In addition, more objective and precise identification was performed using rDNA sequence analysis. The survey identified 286 species that were classified into 2 phyla, 6 classes, 18 orders, 75 families, and 167 genera. Among these, 17 species�Lepiota ignivolvata Bousset & Joss. ex Joss., 1990; Entoloma sericeum Quél., 1872; Hygrophorus queletii Bres., 1881; Inocybe albodiscoides Matheny, 2022; Flammulina rossica Redhead & R. H. Petersen, 1999; Homophron helvolescens (S. Imai) Beker & U. Eberh, 2022; Stropharia lignicola E. J. Tian, 2021; Tengioboletus subglutinosus Yang Wang, Bo Zhang & Yu Li, 2022; Ramaria gracilis (Pers.) Quél., 1888; Hymenochaete huangshanensis S.H. He & Y.C. Dai, 2012; Dacryobolus angiospermarum S.H. He, 2018; Physisporinus eminens (Y.C. Dai) F. Wu, Jia J. Chen & Y.C. Dai, 2017; Cyanosporus bifarius (Spirin) B.K. Cui & Shun Liu, 2021; Fuscopostia leucomallella (Murrill) B.K. Cui, L.L. Shen & Y.C. Dai, 2018; Lactarius fulvihirtipes X.H. Wang, 2018; Russula albolutea B. Chen & J.F. Liang, 2021; and Russula cremicolor G.J. Li & C.Y. Deng, 2020�were recorded for the first time in Korea. Microscopic measurements and images of the 17 species were obtained using an Olympus CX43 microscope (Olympus, Tokyo, Japan). Twenty randomly selected mature basidiospores and basidia from each specimen were evaluated and compared to previously published descriptions [1-14]. Taxonomic classification of species and associated nomenclature were performed using the MycoBank database (http://www.mycobank.org/). For molecular identification, total DNA was extracted from the dried specimens using an AccuPrep Genomic DNA Extraction Kit (Bioneer, Daejeon, Korea). The nuclear large subunit rDNA D1–D2 domains (28S) of Tengioboletus subglutinosus were amplified using primer sets LR0R/LR5 [15,16]. The ITS regions of the other 16 species were amplified using primer sets ITS1F and ITS4B [16]. PCR amplification was performed as previously described [17]. Twenty-four nucleotide sequences of these 17 species have been deposited in GenBank (accession numbers: OR565247–OR565251, OR565254–OR565271, OR54006) and compared with GenBank reference sequences (Table 1). We used MEGA 7 to assemble, proofread, and edit the DNA sequences [18]. The sequences obtained in this study were compared with reference sequences from GenBank A neighbor-joining phylogenetic analysis was performed using MEGA 7 software with Kimura 2-parameter correction. The robustness of the inferred neighbor-joining topologies was tested using 1000 bootstrap replicates (Fig. 1). Using a combination of morphological and phylogenetic analyses, one specimen (NIBRFG0000513774) formed a monophyletic clade with the reference sequence Lepiota ignivolvata (bootstrap=100%; sequence similarity: 100%). One specimen (NIBRFG0000509002) formed a monophyletic clade with the reference sequence of Entoloma sericeum (bootstrap=86%; sequence similarity: 99.7%). Two specimens (NIBRFG0000513807 and NIBRFG0000513813) formed a monophyletic clade with the reference sequences of Hygrophorus queletii (bootstrap=100%; sequence similarity: 100%). One specimen (NIBRFG0000513812) formed a monophyletic clade with the reference sequence of Inocybe albodiscoides (bootstrap=99%; sequence similarity: 99.3%). Two specimens (NIBRFG0000513790 and NIBRFG0000514550) formed a monophyletic clade with the reference sequences of Flammulina rossica (bootstrap support=97%; sequence similarity: 99.7-99.9%). One specimen (NIBRFG0000508845) formed a monophyletic clade with the reference sequence of Homophron helvolescens (bootstrap support=95%; sequence similarity: 99.9%). Two specimens (NIBRFG0000509011 and NIBRFG0000513712) formed a monophyletic clade with the reference sequences of Stropharia lignicola (bootstrap support=100%; sequence similarity: 100%). One specimen (NIBRFG0000513708) formed a monophyletic clade with the reference sequence of Tengioboletus subglutinosus (bootstrap support=100%; sequence similarity: 100%). Two specimens (NIBRFG0000508844 and NIBRFG0000513701) formed a monophyletic clade with the reference sequences of Ramaria gracilis (bootstrap support=100%; sequence similarity: 100%). One specimen (NIBRFG0000511214) formed a monophyletic clade with the reference sequence of Hymenochaete huangshanensis (bootstrap support=100%; sequence similarity: 100%). Two specimens (NIBRFG0000505734 and NIBRFG0000510350) formed a monophyletic clade with the reference sequences of Dacryobolus angiospermarum (bootstrap support=100%; sequence similarity: 99.8100%). One specimen (NIBRFG0000511205) formed a monophyletic clade with the reference sequence of Physisporinus eminens (bootstrap support=100%; sequence similarity: 99.7%). One specimen (NIBRFG0000513779) formed a monophyletic clade with the reference sequence of Cyanosporus bifarius (bootstrap support=84%; sequence similarity: 99.2%). One specimen (NIBRFG0000513753) formed a monophyletic clade with the reference sequences of Fuscopostia leucomallella (bootstrap support=100%; sequence similarity: 99.5%). Three specimens (NIBRFG0000508975, NIBRFG0000508989, and NIBRFG0000509020) formed a monophyletic clade with the reference sequences of Lactarius fulvihirtipes (bootstrap support=87%; sequence similarity: 99.4%). One specimen (NIBRFG0000508947) formed a monophyletic clade with the reference sequence of Russula albolutea (bootstrap support=100%; sequence similarity: 99.5%). One specimen (NIBRFG000051121) formed a monophyletic clade with the reference sequence of Russula cremicolor (bootstrap support=100%; sequence similarity: 100%). Here, 17 newly confirmed unrecorded species were added to the Korean National Species List [19]. Photographs of fruiting bodies, drawings of microscopic features (Fig. 1), and descriptions and discussions of these species are presented.
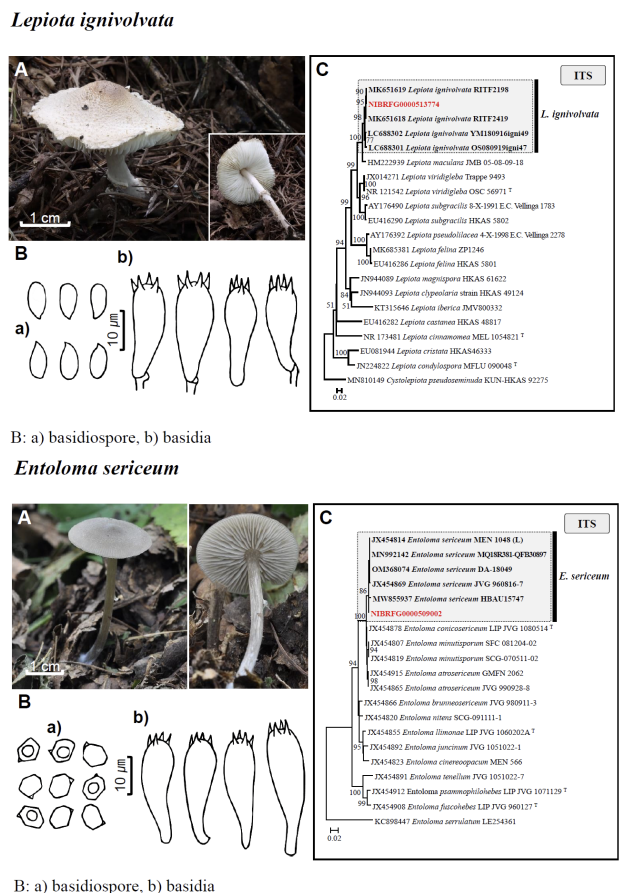
Fig. 1 Fruiting bodies (A), microscopic features (B), and neighbor-joining tree (C) of 17 unrecorded species: Lepiota ignivolvata, Entoloma sericeum, Hygrophorus queletii, Inocybe albodiscoides, Flammulina rossica, Homophron helvolescens, Stropharia lignicola, Tengioboletus subglutinosus, Ramaria gracilis, Hymenochaete huangshanensis, Dacryobolus angiospermarum, Physisporinus eminens, Cyanosporus bifarius, Fuscopostia leucomallella, Lactarius fulvihirtipes, Russula albolutea, and Russula cremicolor. (C) Bootstrap scores > 50 are shown at the nodes. The scale bar indicates the number of nucleotide substitutions per site. ITS, internal transcribed spacer; LSU, large subunit. (continued)
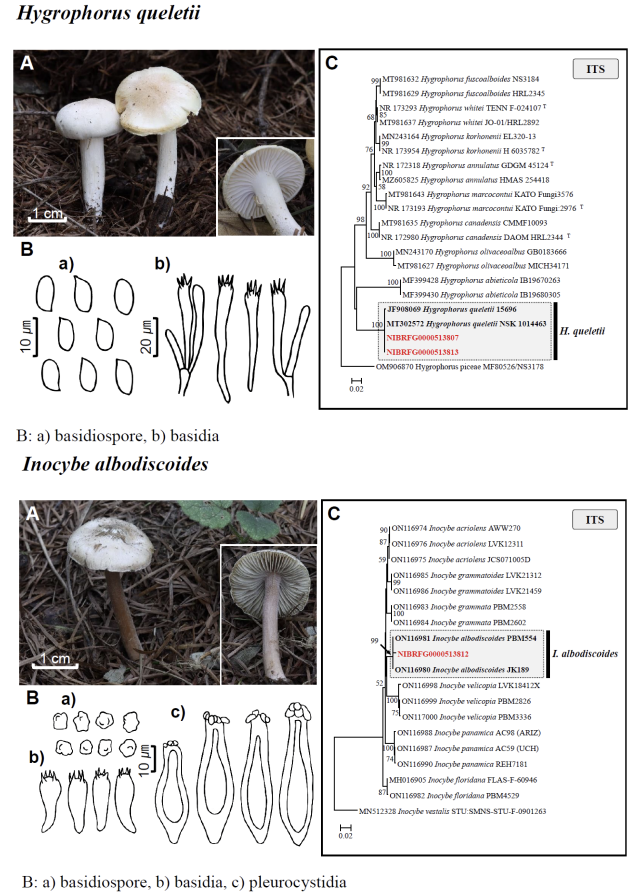
Fig. 1 Fruiting bodies (A), microscopic features (B), and neighbor-joining tree (C) of 17 unrecorded species: Lepiota ignivolvata, Entoloma sericeum, Hygrophorus queletii, Inocybe albodiscoides, Flammulina rossica, Homophron helvolescens, Stropharia lignicola, Tengioboletus subglutinosus, Ramaria gracilis, Hymenochaete huangshanensis, Dacryobolus angiospermarum, Physisporinus eminens, Cyanosporus bifarius, Fuscopostia leucomallella, Lactarius fulvihirtipes, Russula albolutea, and Russula cremicolor. (C) Bootstrap scores > 50 are shown at the nodes. The scale bar indicates the number of nucleotide substitutions per site. ITS, internal transcribed spacer; LSU, large subunit. (continued)
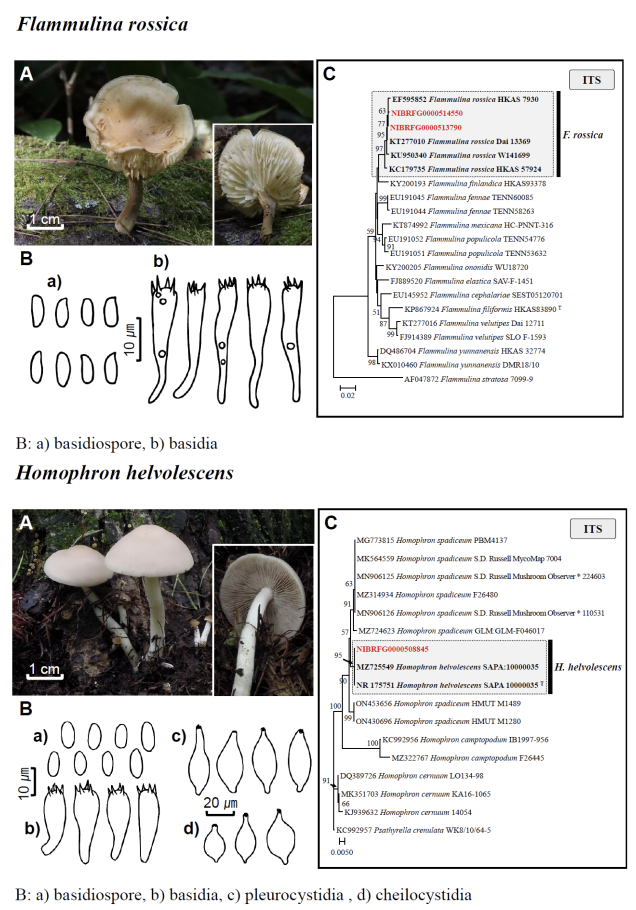
Fig. 1 Fruiting bodies (A), microscopic features (B), and neighbor-joining tree (C) of 17 unrecorded species: Lepiota ignivolvata, Entoloma sericeum, Hygrophorus queletii, Inocybe albodiscoides, Flammulina rossica, Homophron helvolescens, Stropharia lignicola, Tengioboletus subglutinosus, Ramaria gracilis, Hymenochaete huangshanensis, Dacryobolus angiospermarum, Physisporinus eminens, Cyanosporus bifarius, Fuscopostia leucomallella, Lactarius fulvihirtipes, Russula albolutea, and Russula cremicolor. (C) Bootstrap scores > 50 are shown at the nodes. The scale bar indicates the number of nucleotide substitutions per site. ITS, internal transcribed spacer; LSU, large subunit. (continued)
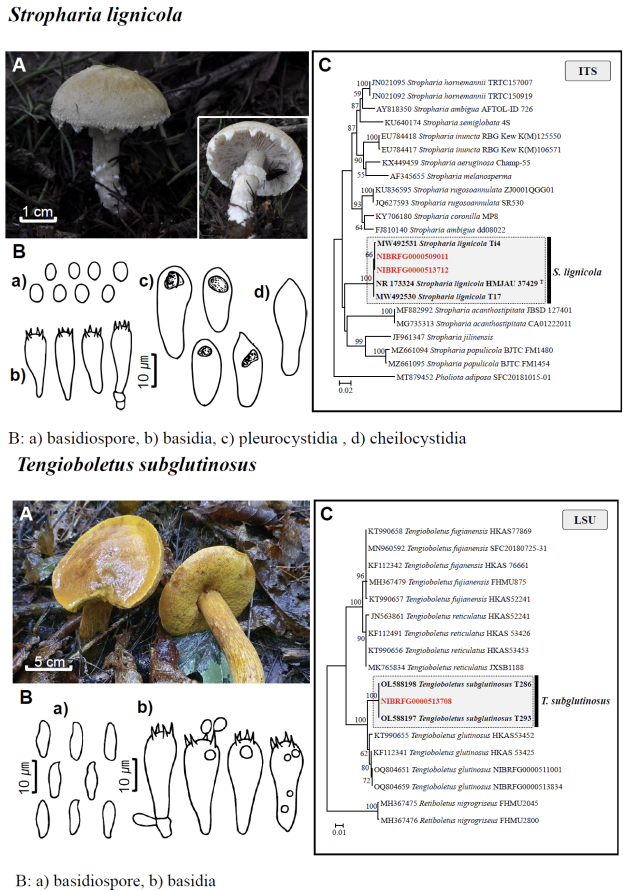
Fig. 1 Fruiting bodies (A), microscopic features (B), and neighbor-joining tree (C) of 17 unrecorded species: Lepiota ignivolvata, Entoloma sericeum, Hygrophorus queletii, Inocybe albodiscoides, Flammulina rossica, Homophron helvolescens, Stropharia lignicola, Tengioboletus subglutinosus, Ramaria gracilis, Hymenochaete huangshanensis, Dacryobolus angiospermarum, Physisporinus eminens, Cyanosporus bifarius, Fuscopostia leucomallella, Lactarius fulvihirtipes, Russula albolutea, and Russula cremicolor. (C) Bootstrap scores > 50 are shown at the nodes. The scale bar indicates the number of nucleotide substitutions per site. ITS, internal transcribed spacer; LSU, large subunit. (continued)
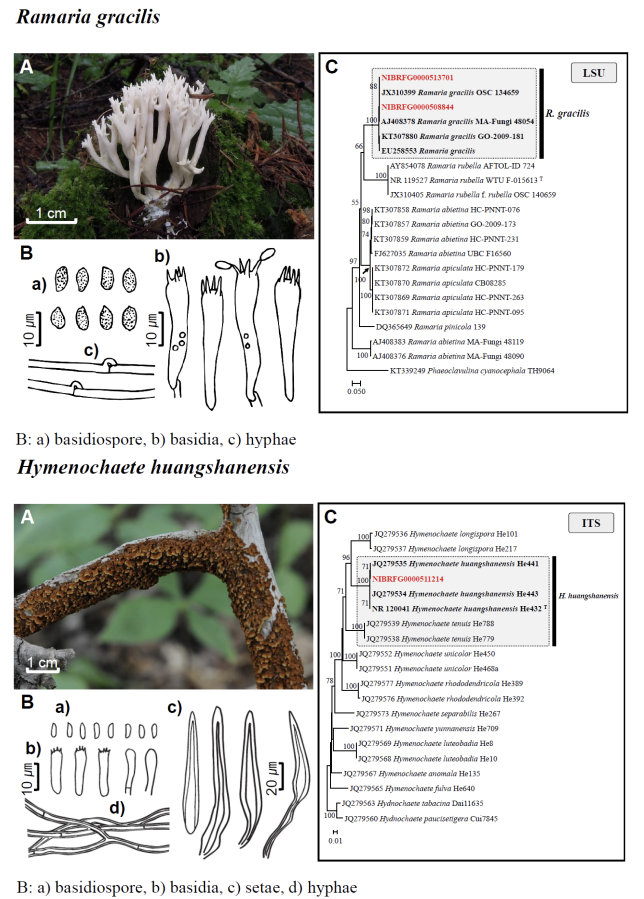
Fig. 1 Fruiting bodies (A), microscopic features (B), and neighbor-joining tree (C) of 17 unrecorded species: Lepiota ignivolvata, Entoloma sericeum, Hygrophorus queletii, Inocybe albodiscoides, Flammulina rossica, Homophron helvolescens, Stropharia lignicola, Tengioboletus subglutinosus, Ramaria gracilis, Hymenochaete huangshanensis, Dacryobolus angiospermarum, Physisporinus eminens, Cyanosporus bifarius, Fuscopostia leucomallella, Lactarius fulvihirtipes, Russula albolutea, and Russula cremicolor. (C) Bootstrap scores > 50 are shown at the nodes. The scale bar indicates the number of nucleotide substitutions per site. ITS, internal transcribed spacer; LSU, large subunit. (continued)
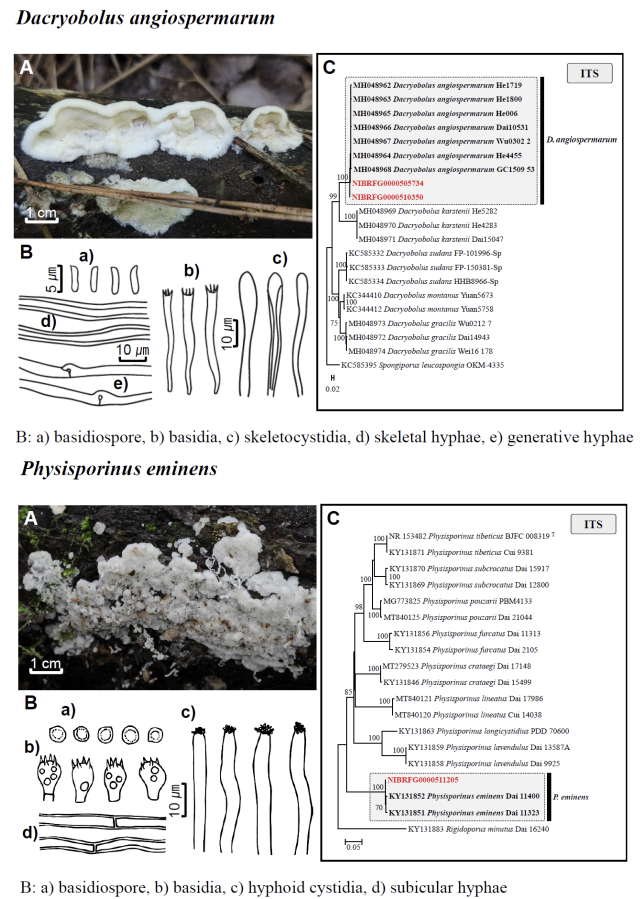
Fig. 1 Fruiting bodies (A), microscopic features (B), and neighbor-joining tree (C) of 17 unrecorded species: Lepiota ignivolvata, Entoloma sericeum, Hygrophorus queletii, Inocybe albodiscoides, Flammulina rossica, Homophron helvolescens, Stropharia lignicola, Tengioboletus subglutinosus, Ramaria gracilis, Hymenochaete huangshanensis, Dacryobolus angiospermarum, Physisporinus eminens, Cyanosporus bifarius, Fuscopostia leucomallella, Lactarius fulvihirtipes, Russula albolutea, and Russula cremicolor. (C) Bootstrap scores > 50 are shown at the nodes. The scale bar indicates the number of nucleotide substitutions per site. ITS, internal transcribed spacer; LSU, large subunit. (continued)
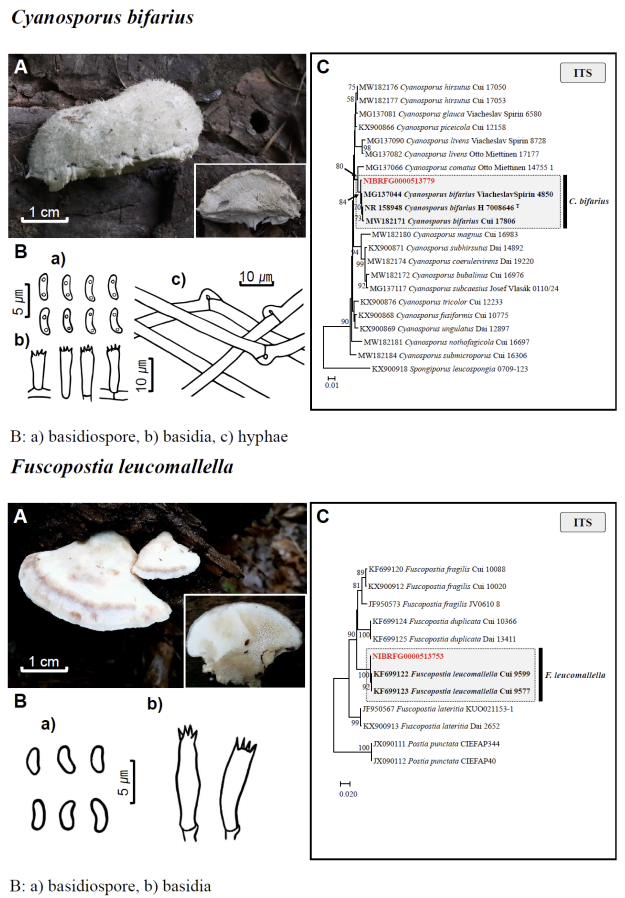
Fig. 1 Fruiting bodies (A), microscopic features (B), and neighbor-joining tree (C) of 17 unrecorded species: Lepiota ignivolvata, Entoloma sericeum, Hygrophorus queletii, Inocybe albodiscoides, Flammulina rossica, Homophron helvolescens, Stropharia lignicola, Tengioboletus subglutinosus, Ramaria gracilis, Hymenochaete huangshanensis, Dacryobolus angiospermarum, Physisporinus eminens, Cyanosporus bifarius, Fuscopostia leucomallella, Lactarius fulvihirtipes, Russula albolutea, and Russula cremicolor. (C) Bootstrap scores > 50 are shown at the nodes. The scale bar indicates the number of nucleotide substitutions per site. ITS, internal transcribed spacer; LSU, large subunit. (continued)
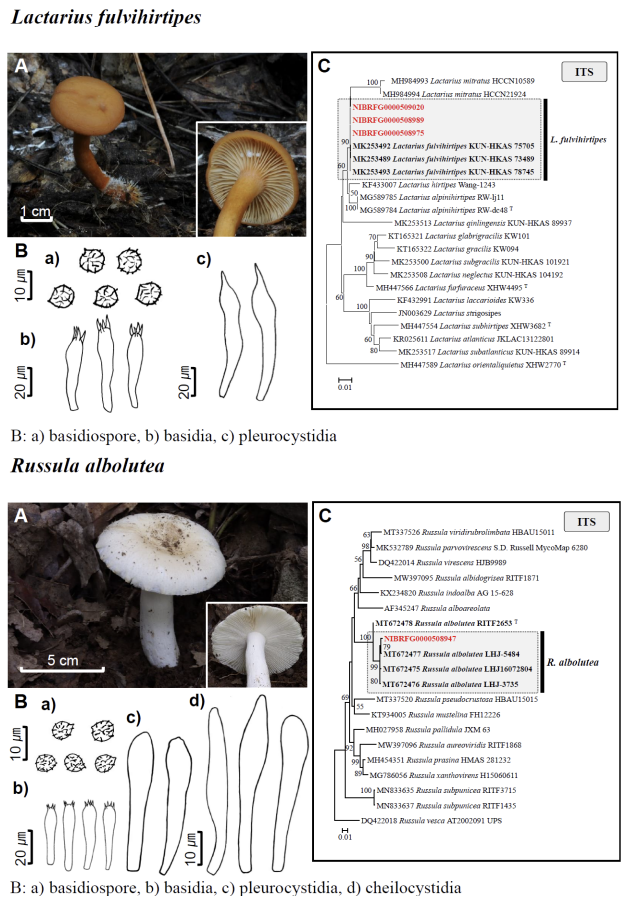
Fig. 1 Fruiting bodies (A), microscopic features (B), and neighbor-joining tree (C) of 17 unrecorded species: Lepiota ignivolvata, Entoloma sericeum, Hygrophorus queletii, Inocybe albodiscoides, Flammulina rossica, Homophron helvolescens, Stropharia lignicola, Tengioboletus subglutinosus, Ramaria gracilis, Hymenochaete huangshanensis, Dacryobolus angiospermarum, Physisporinus eminens, Cyanosporus bifarius, Fuscopostia leucomallella, Lactarius fulvihirtipes, Russula albolutea, and Russula cremicolor. (C) Bootstrap scores > 50 are shown at the nodes. The scale bar indicates the number of nucleotide substitutions per site. ITS, internal transcribed spacer; LSU, large subunit. (continued)
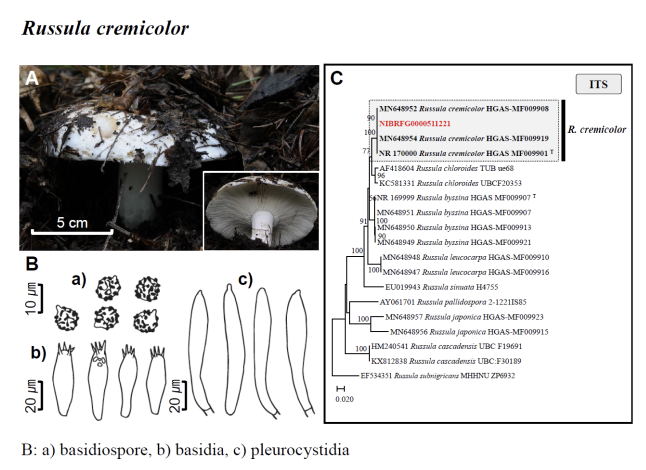
Fig. 1 Fruiting bodies (A), microscopic features (B), and neighbor-joining tree (C) of 17 unrecorded species: Lepiota ignivolvata, Entoloma sericeum, Hygrophorus queletii, Inocybe albodiscoides, Flammulina rossica, Homophron helvolescens, Stropharia lignicola, Tengioboletus subglutinosus, Ramaria gracilis, Hymenochaete huangshanensis, Dacryobolus angiospermarum, Physisporinus eminens, Cyanosporus bifarius, Fuscopostia leucomallella, Lactarius fulvihirtipes, Russula albolutea, and Russula cremicolor. (C) Bootstrap scores > 50 are shown at the nodes. The scale bar indicates the number of nucleotide substitutions per site. ITS, internal transcribed spacer; LSU, large subunit. (continued)
Table 1 Closest GenBank matches of 17 undescribed species in this study.
TAXONOMY
Basidiomycota R.T. Moore
Agaricomycetes Doweld
Agaricales Underw.
Agaricaceae Chevall.
Homophron (Britzelm.) Örstadius & E. Larss., 2015
Korean name. Mu-Reun-Ju-Reum-Beo-Seot-Sok, nom. nov. (무른주름버섯속)
Type: Homophron spadiceum (P. Kumm.) Örstadius & E. Larss. 2015
Boletales E.-J. Gilbert
Boletaceae Chevall.
Gomphales Jülich
Gomphaceae Donk
Hymenochaetales Oberw
Hymenochaetaceae Donk
Polyporales Gäum.
Dacryobolaceae Jülich
Meripilaceae Jülich
Physisporinus P. Karst., 1889
Korean name. Ggeop-Jil-Gu-Meong-Beo-Seot-Sok, nom. nov. (껍질구멍버섯속)
Type: Physisporinus vitreus (Pers.) P. Karst., 1889
Polyporaceae Fr. ex Corda
Cyanosporus McGinty, 1909
Korean name. Pu-Reun-Sal-Beo-Seot-Sok, nom. nov. (푸른살버섯속)
Type: Cyanosporus caesius (Schrad.) McGinty, 1909
Postiaceae B.K. Cui, Shun Liu & Y.C. Dai
Fuscopostia B.K. Cui, L.L. Shen & Y.C. Dai, 2018
Korean name. Gal-Saek-Son-Deung-Beo-Seot-Sok, nom. nov. (갈색손등버섯속)
Type: Fuscopostia fragilis (Fr.) B.K. Cui, L.L. Shen & Y.C. Dai, 2018
Russulales Kreisel ex P.M. Kirk, P.F. Cannon & J.C. Davi
Russulaceae Lotsy
