Introduction
Microbiome research of soil invertebrates have received much attention [1,2]. Recently, a study reported that terrestrial insects, such as collembolans, earthworms, and nematodes, are associated with an abundant microbiome and putative symbionts [3]. Earthworms are excellent bioindicators of soil quality and perform an important role in increasing soil aeration, infiltration, structure, nutrient cycling, water movement, and plant growth [4].
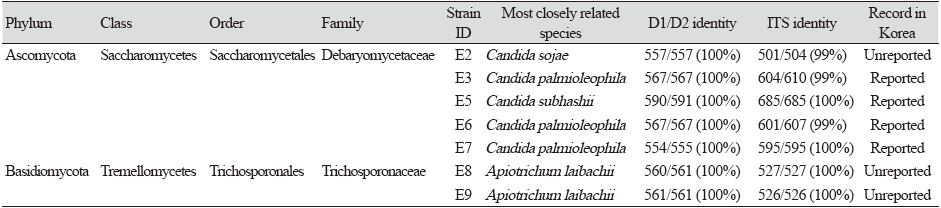
In 2021, seven strains were isolated from earthworm (Eisenia andrei) gut samples collected from the Nanji Water Regeneration Center in Korea. Among the seven strains, five were Ascomycota yeast and two were Basidiomycota yeast. The five strains were assigned to the genus Candida of the order Saccharomycetales of class Saccharomycetes in the phylum Ascomycota, and two were assigned to the genus Apiotrichum (two strains) of the order Trichosporonales of the class Tremellomycetes in the phylum Basidiomycota. Three of the isolated strains; E2, E8, and E9, have not previously been reported in Korea.
The genus Candida consists of 314 recognized species and has C. vulgaris [5] as a type species. Candida species have been isolated from human or animal excrement and aquatic environments containing urban sewage effluent [6-8], flowers, and insects [9-12]. The cells are globose, ellipsoidal, cylindroidal, or elongate, sometimes with ogival, triangular, or lunate shape, and reproduce through holoblastic budding [5].
The genus Apiotrichum consists of 22 reported species [13,14] and has Apiotrichum (= Trichosporon) porosum [15] as the type species. The genus Apiotrichum was redefined to accommodate gracile/brassicae [15-18]. Yeasts of this genus are found distributed in nature, on sources such as cabbage, rotten wood, sour milk, and grassland [19] and many species have been isolated from the soil [17,18]. Apiotrichum presents pseudohyphae and budding cells, but basidiocarps, sexual reproduction, fermentation, and nitration [15,17].
Materials and methods
The host worms were collected from the Nanji Water Regeneration Center in Goyang City, Gyeonggi Province, Korea, and a total of seven strains were isolated from the intestinal tract of the worms. The intestine was separated from E. andrei, cut to approximately 2 cm with tweezers, pulverized, and washed continuously with distilled water [20]. The suspension containing the microbes was spread (100 µL each) on yeast mold agar (YM agar; Difco, Franklin Lakes, USA). Single yeast colonies were purified and maintained on YM agar containing 25% (w/v) glycerol suspension at -80℃ in a deep freezer [21,22]. Information on the designated strain identifications (IDs), recent associations, 26S rRNA similarities, and internal transcribed spacer (ITS) similarities for isolated strains are described in Table 1.
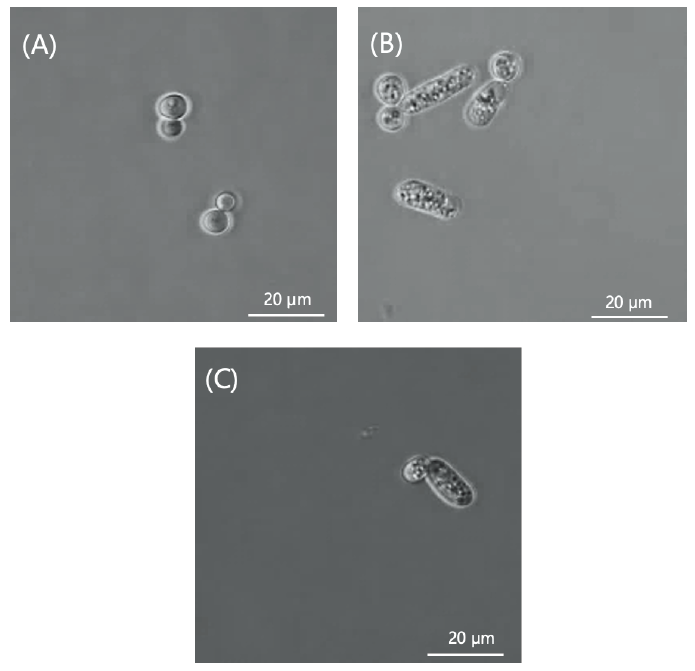
The incubation period is generally 23 days at 25℃, but the incubation temperature and times range between 4-42℃ and 3 days or more, respectively, for each strain [23]. Growth tests were performed on different media, such as YM agar, corn meal agar (CMA; Difco, Franklin Lakes, USA), and potato dextrose agar (PDA; Difco, Franklin Lakes, USA), at 10℃ (Fig. 1). Budding and cell morphology were observed using a phase-contrast microscope (Leica DM500, Wetzlar, Germany), using pure cultured cells incubated for 3-5 days in YM agar. Phase contrast microscope images and pictures of plates containing the strains are shown in Fig. 2. The API 20C AUX kit (BioMérieux, Marcy-l'Étoile, France) was used to determine the carbon source assimilation of the unrecorded yeast strains.
The yeast used for DNA extraction was subcultured on YM agar and incubated at 10℃ for 3-5 days. Genomic DNA was extracted using a cDNA Synthesis Kit (NanoHelix, Daejeon, Korea) according to the manufacturer's instructions. For strain classification, the sequences were analyzed by amplifying the D1/D2 region of the large subunit (LSU) RNA gene using the universal primers NL1 (5'-GCATCAAGGAGAG-3') and NL4 (5'-GTCGTTCAGG-3') [24]. The types of yeast closely associated with the isolated strains were collected using the MYCOBANK database (https://www.mycobank.org/). The LSU rDNA sequences of the strains were obtained from NCBI (https://www.ncbi.nlm.nih.gov/), and homology with other yeast was compared using the Basic Local Alignment Search Tool (BLAST) database of NCBI. The gene sequences were edited using the Seqman program and used for constructing phylogenetic trees using the neighbor-joining algorithm [25] of the MEGA7 program [26]. The phylogenetic tree topology was evaluated for statistical reliability based on bootstrap values for 1,000 replications [27] and using the GenBank registration number. The evolutionary distance was calculated using the Kimura 2-parameter model [28].
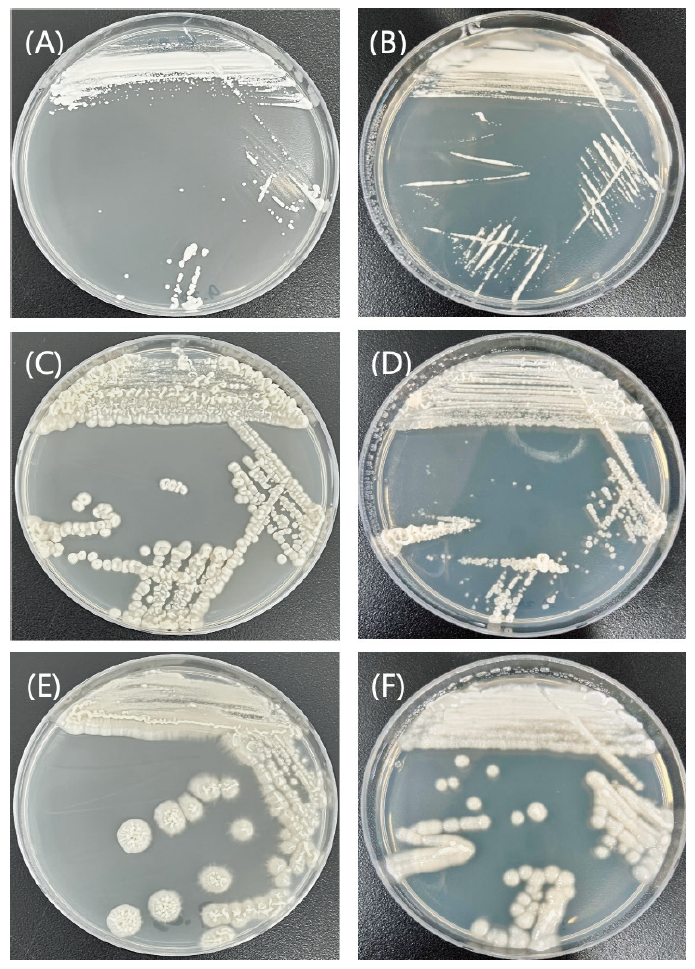
Fig. 1. Morphology of cells from the unrecorded strains incubated at 10℃. The colonies of (A) Candida sojae E2, (C) Apiotrichum laibachii E8, and (E) A. laibachii E9 cultured in yeast mold agar (YM). The colonies of (B) C. sojae E2, (D) A. laibachii E8, and (F) A. laibachii E9 cultured in potato dextrose agar (PDA).
Results and Discussion
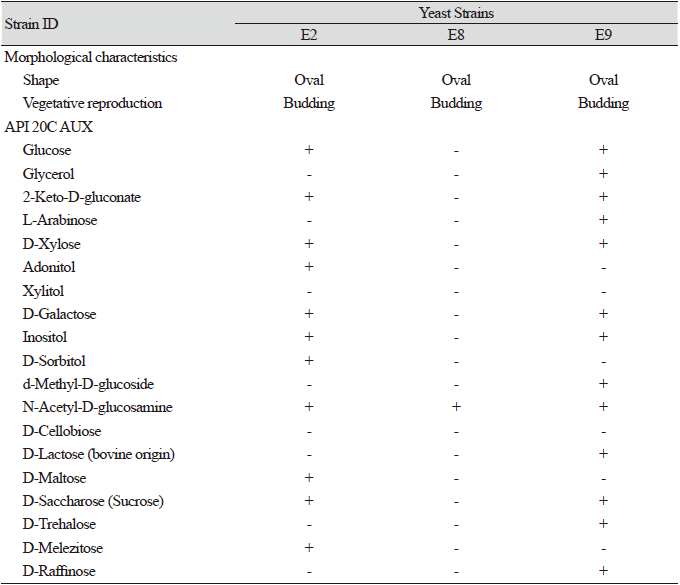
A total of seven strains of yeast were separated from the Nanji Water Regeneration Center in Gyeonggi Province, Korea. By analyzing the similarities in the small subunit (SSU) rDNA and D1/D2 region of LSU rDNA sequence, we found that three of the separated strains were unrecorded in Korea. There was a higher proportion of Ascomycota yeast among the strains isolated from the internal contents of the E. andrei, compared to Basidiomycota yeast. Five of the seven strains were identified as belonging to the family Debaryomycetaceae of the phylum Ascomycota, while the remaining two strains were classified into Trichosporonaceae of the phylum Basidiomycota. The D1/D2 and ITS similarities between the strains and their closely related species and their taxonomic composition are summarized in Table 1. The characteristics of the unrecorded strains are presented in Table 2.
Table 2. Characteristics of the unrecorded strains from worm gut.
|
|
All data were obtained in this study. +, positive; w, weakly positive; -, negative. |
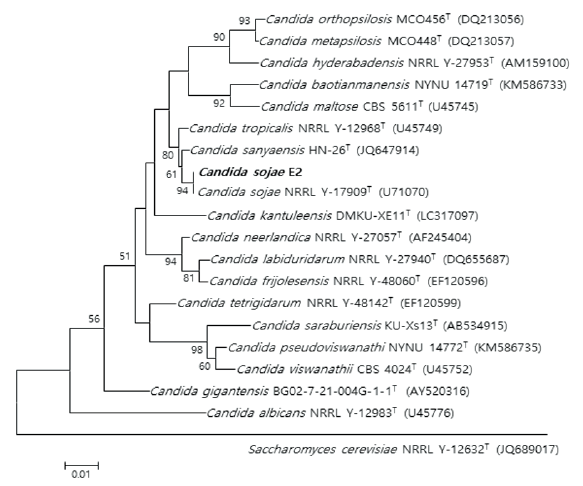
Fig. 3. Neighbor-joining phylogenetic tree based on 26S rRNA gene sequences shows the phylogenetic relationships between the strain E2 and their closest strains of the genus Candida. Saccharomyces cerevisiae NRRL Y-12632T strain was used as the outgroup. Bootstrap values (>50 %) aresh own at each branch. Bar, 0.01 substitutions per nucleotide position. T: Type strain. Bold type font: isolated strain.
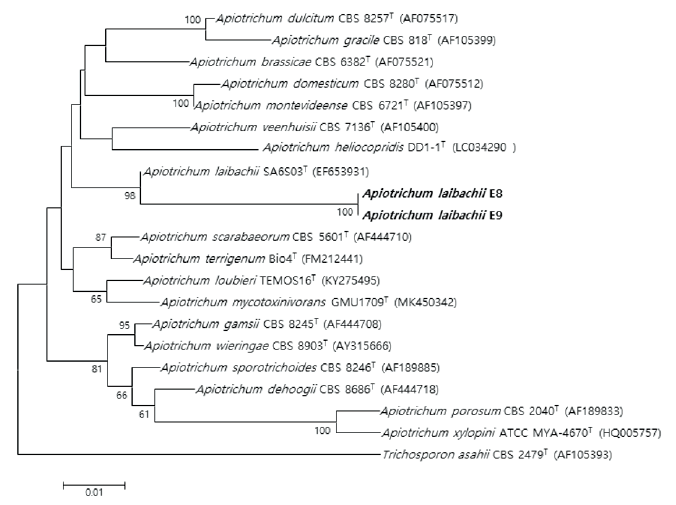
Fig. 4. Neighbor-joining phylogenetic tree based on 26S rRNA gene sequences shows the phylogenetic relationships between the strains E8, E9 and their closest strains of the genus Apiotrichum. Trichosporon asahii CBS 2479T strain was used as the outgroup. Bootstrap values (>50 %) are shown at each branch. Bar, 0.01 substitutions per nucleotide position. T: Type strain. Bold type font: isolated strain.
Three unrecorded yeasts were identified: Candida (1 strain) and Apiotrichum (2 strains). The phylogenetic consensus trees of the three strains support a close relationship by demonstrating that the isolated strains are closely linked to strains that exhibit the highest LSU rDNA sequence similarity (Fig. 3 and 4).
Based on previous phylogenetic and biochemical studies, this study identified three unrecorded yeasts in the domestic ecosystem and investigated their phenotypic characteristics.
Description of Candida sojae E2
Colonies are convex, entire, and cream-colored after 3 days of incubation on YM agar and PDA at 10℃. The strain grew on YM, PDA, and CMA (weakly) (Fig. 1). In the API 20C AUX test, strain E2 was positive for D-sorbitol, inositol, glucose, D-melezitose, D-saccharose (sucrose), D-maltose, D-galactose, D-xylose, adonitol, N-acetyl-D-glucosamine, and 2-keto-D-gluconate. However, it was negative for D-raffinose, D-trehalose, D-cellobiose, d-methyl-D-glucoside, xylitol, glycerol, L-arabinose, and D-lactose (bovine origin). Strain E2 (KCTC 27832) was isolated from the intestinal tract of the earthworms obtained from the Nanji Water Regeneration Center, Gyeonggi Province, Korea.
Description of Apiotrichum laibachii E8
Colonies were entire, umbonate, and cream-colored after 3 days of incubation on YM agar at 10℃ and observed to form an avillous appearance in which the center protrudes. The strain grew on YM, PDA, and CMA (weakly) (Fig. 1). In the API 20C AUX test, strain E8 was positive for N-acetyl-D-glucosamine. However, it was negative for glucose, glycerol, 2-keto-D-gluconate, L-arabinose, D-sorbitol, d-methyl-D-glucoside, D-cellobiose, D-lactose (bovine origin), D-maltose, D-saccharose (sucrose), D-trehalose, D-melezitose, xylitol, D-galactose, adonitol, inositol, and D-raffinose. Strain E8 (KCTC 27842) was isolated from the intestinal tract of the earthworms obtained from the Nanji Water Regeneration Center, Gyeonggi Province, Korea.
Description of Apiotrichum laibachii E9
Colonies were entire, umbonate, and cream-colored after 3 days of incubation on YM agar and PDA at 10℃ and observed to form an avillous appearance in which the center protrudes. The strain grew on YM, PDA, and CMA (weakly) (Fig. 1). In the API 20C AUX test, strain E9 was positive for D-lactose (bovine origin), N-acetyl-D-glucosamine, d-methyl-D-glucoside, D-galactose, glucose, 2-keto-D-gluconate, L-arabinose, D-xylose, D-saccharose (sucrose), D-trehalose, and D-raffinose. However, it was negative for D-melezitose, D-cellobiose, xylitol, adonitol, D-maltose, D-sorbitol, glycerol, and inositol. Strain E9 (KCTC 27843) was isolated from the intestinal tract of the earthworms from the Nanji Water Regeneration Center, Gyeonggi Province, Korea.