INTRODUCTION
The class Sordariomycetes includes taxa with perithecial ascomata, paraphysate hamathecium, periphysate ostioles, and unitunicate or pseudoprotunicate asci [1]. Sordariomycetes is the second largest class of Ascomycota and contains 1,331 genera and 105 families, including 32 orders and 6 subclasses [2]. The genus Diaporthe, belonging to the class Sordariomycetes, was identified by Nitschke, using the type species D. eres from Ulmus sp. collected in Germany. It belongs to the family Diaporthaceae and order Diaporthales [2,3]. Numerous species names in the genus Diaporthe have emerged over the years because of their introduction, which is mostly based on host relationships. A single species can be found in one or more hosts, and it is now understood that the species are not host-specific [4]. The key factor used to classify different Diaporthe species is their molecular phylogeny, particularly those based on the internal transcribed spacer (ITS) of nuclear ribosomal DNA. However, some authors have also used the translation elongation factor 1-alpha (TEF1α) gene [5]. The classification of the genus relies heavily on molecular phylogenies, particularly those generated from sequences of the ITS, TEF1α, β-tubulin (TUB2), and calmodulin (CAL) loci [6]. Another member of the class Sordariomycetes, belonging to the genus Faurelina was first described as a new genus of cleistothecial ascomycetes [7]. This genus has a problematic taxonomic history. Initially, Faurelina was tentatively placed in Chadefaudiellaceae; however, this position was questionable. The type species Faurelina belongs to the class of ascolocular ascomycetes and should be placed in the class formerly known as Loculoascomycetes [8]. Faurelina was then transferred to Microascaceae after two years, mainly based on the presence of dextrinoid ascospores without germ pores [9]. The placement of Faurelina in Microascales partly resulted from the study by [10], who sequenced the nucLSU, nucSSU, and RPB2 gene regions from a single strain labeled F. indica (CBS 126.78), giving results identical to those of the type species Ceratocystis fimbriata.
In addition, the well-known genus belonging to the class Sordariomycetes, called Trichoderma, is distinguished by perithecia submerged in fleshy stromata of various colors, cylindrical asci with green or hyaline ascospores that disarticulate at the septum, conidiophores that create various branch patterns, and green or hyaline conidia [11]. The taxonomic classification of Trichoderma spp. using traditional methods is challenging. Recently, the study of genus taxonomy has advanced based on combined assessments of morphology and DNA sequence data, demonstrating that the color of ascospores can reveal important evolutionary aspects [12]. Although Trichoderma is a well-studied genus, there are several species whose evolutionary positions are not fully understood [13].
The identification and classification of fungal species isolated from Korean soil was the main goal of this extensive investigation. Fungal species, together with their morphological and cultural traits, were identified using molecular phylogenetic analyses. In this study, three fungal species from Korea are described and illustrated as new records.
MATERIAL AND METHODS
Soil sampling and fungal isolation
In 2018 and 2019, soil samples were collected from Jeju Island (33°25'22.9"N, 126°33'01.6"E) and Gyeongsangbuk-do (35°51'39.7"N, 129°16'09.5"E), South Korea. Using a sterile pre-autoclaved spatula, soils from depths of 10-30 cm were randomly sampled, transferred to sterile zipper plastic bags, and then transported to the laboratory. The soil samples (1 g) were suspended in 10 mL of sterile distilled water, vortexed gently, and serially diluted, and a volume of 100 μL was spread on potato dextrose agar (PDA; Difco, Detroit, MI, USA) plates and incubated for 2-3 days at 25℃. The germinating single colonies were then transferred to new PDA plates at 25℃. A pure culture of the strain was selected for further molecular analysis based on specific characteristics. For further study, the fungal strains were maintained in 20% glycerol at -80℃. Stock cultures of three strains, namely KNU-JJ-1830 (NIBRFGC000502239), KNU-JJ-1809 (NIBRFGC000502235), and KNU-4-KH1 (NIBRFGC000505959), were deposited at the National Institute of Biological Resources (NIBR) as metabolically inactive cultures.
Cultural and morphological characterization
Cultural and morphological characteristics were evaluated using different media, depending on the genus and species. KNU-JJ-1809 and KNU-JJ-1830 were cultured on PDA, oatmeal agar (OA; Difco, Detroit, MI, USA), and malt extract agar (MEA; Difco, Detroit, MI, USA) and incubated at 25℃ for 2 and 3 weeks, respectively [6,14]. KNU-4-KH1 was transferred on PDA, MEA, cornmeal dextrose agar (CMAD: cornmeal, 40.0 g/L; dextrose, 20.0 g/L; agar, 15.0 g/L; distilled water, 1,000 mL), and synthetic nutrient agar (SNA) followed by 2 weeks at 25℃ [15,16]. Next, cultural features such as colony color, fungal development, and texture, were documented. Mycological characteristics of the fungal strains were observed under a light microscope (BX-50, Olympus, Tokyo, Japan).
Genomic DNA extraction, PCR amplification, and sequencing
Total genomic DNA was extracted using the HiGene Genomic DNA Prep Kit (BIOFACT, Daejeon, Korea) according to the manufacturer’s instructions. The PCR amplification process was conducted using a fragment of the ITS regions as a target using primers ITS1F/ITS4 [17,18]; 28S ribosomal DNA of large subunit using LROR/LR5 [19]; TEF1α (EF1-728F/TEF1rev or EF1-986R) [20,21]; CAL (CAL-228F/CAL-737R) [22]; and TUB2 (T1/Bt-2b) [22,23]. The products were run on an ethidium bromide-stained 1% agarose gel to confirm their yields, purified using EXOSAP-IT (Thermo Fisher Scientific, Waltham, MA, USA), and sequenced by Solgent (Daejeon, Korea).
Molecular phylogenetic analysis
For phylogenetic analyses, the related sequences were retrieved from the National Center for Biotechnology Information (Table 1). The MEGA7.0 program was used to align each gene and combine the sequences to show where the strains fell on the phylogenetic tree. Ambiguous regions were deleted from the alignments, and the evolutionary distance matrices for the neighbor-joining (NJ) algorithm were calculated using Kimura’s two-parameter model [24]. The NJ method was inferred from the tree topology using MEGA7 with bootstrap values based on 1,000 replications [25].
RESULTS AND DISCUSSION
Taxonomy
Diaporthe endophytica R. R. Gomes, C. Glienke & Crous, Persoonia 31:20 (2013) (Fig. 1).
Ecology and Distribution: Most members of this genus have been recorded from different hosts, including endophytic, saprobic, and plant-pathogenic fungi such as the leaves of Acacia retinodes (Australia), branches of Betula alleghaniensis (Canada), Pyrus communis (South Africa), Helianthus annuus (Italy), twigs of Ulmus sp. (Germany), stems and twigs of Toxicodendron vernicifluum (Japan), and soil (Korea). The strain KNU-JJ-1809 was collected from Korean soil.
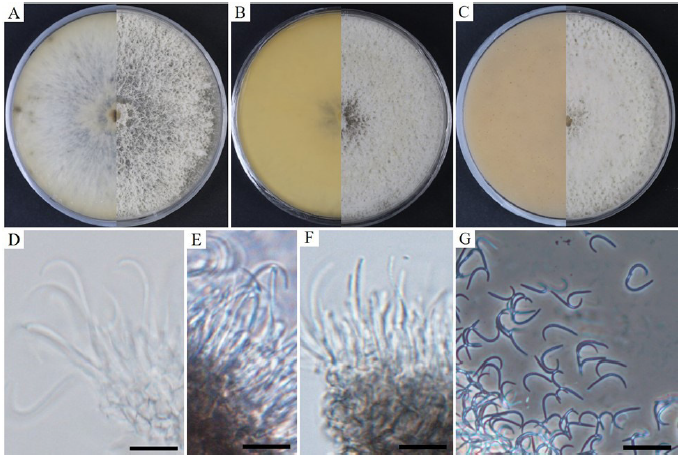
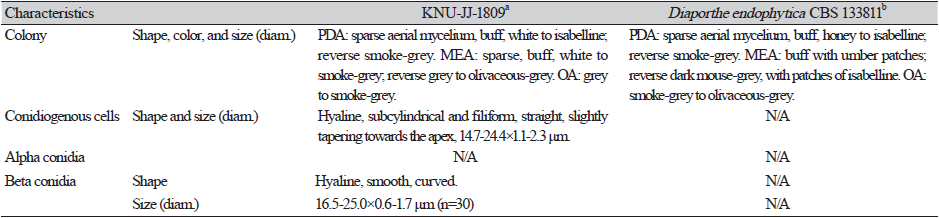
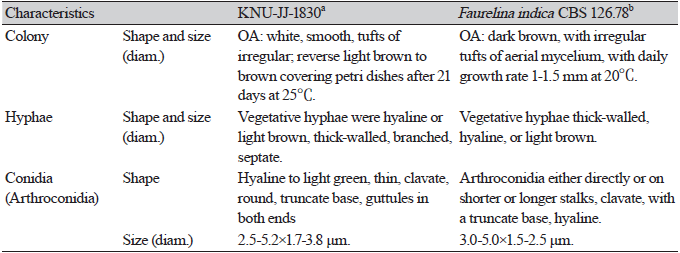
Cultural and morphological characteristics: The strains were cultured on PDA, MEA, and OA media to study their cultural and morphological characteristics (Table 2). The strain produced sparse aerial mycelium, buff, white to isabelline, reverse smoke-grey, covering the petri dish after 2 weeks on PDA at 25℃ (Fig. 1A). The colonies were sparse, buff, white to smoke-grey, reverse grey to olivaceous-grey, and covered the petri dish after 2 weeks on MEA at 25℃ (Fig. 1B) and OA gray to smoke-gray (Fig. 1C). The conidiophores were hyaline, branched, densely aggregated, cylindrical, straight to curved, and had 1–3-septate. Conidiogenous cells were hyaline, subcylindrical and filiform, straight, slightly tapering toward the apex, and with a diameter of (n=10) 14.7-24.4×1.1-2.3 μm (average, 18.7×1.5 μm) (Figs. 1D-F)). No alpha-conidia were observed. Beta conidia were commonly found, straight, curved, hyaline, smooth-walled, and with a diameter (n=30) of 16.5-25.0×0.6-1.7 μm (average=20.1×1.2 μm) (Fig. 1G). Based on the cultural and morphological characteristics, KNU-JJ-1809 was similar to the previously identified D. endophytica (Table 2).
Molecular phylogeny of KNU-JJ-1809
Sequence analysis revealed that 485 bp of the ITS region, 440 bp of CAL, 305 bp of TEF1α, and 445 bp of TUB2 were obtained. The BLAST search results of the ITS sequences revealed that KNU-JJ-1809 exhibited 100% similarity with different strains of D. endophytica (CBS 133811T, NOU760, NTOU 4884, NTOU 4380, NTOU 4920, PHA-1, and ZJUD73). CAL showed maximum 98.2-98.9% similarity with the D. endophytica strains (LGMF937, CBS 133811T, and LGMF935). TEF1α showed maximum 99.3-99.7% similarity with the D. endophytica strains (SoyYS1732, ZJUD94, and ZJUD73), and the TUB2 displayed maximum 99.2% similarity with the D. endophytica strain LGMF935. Phylogenetic analysis was performed based on a combination of ITS, CAL, TEF1α, and TUB2 sequences using the NJ phylogenetic tree. Analysis of the combined sequences in the phylogenetic tree revealed that KNU-JJ-1809 clustered with a previously identified strain of D. endophytica (Fig. 2). Therefore, KNU-JJ-1809 was identified as D. endophytica.
According to previous research, Diaporthe species are common in temperate and tropical regions as endophytes associated with several hosts [26]. D. betulae and D. betuliclola have been identified on the genus Betula in China, and often appear to be the source of birch dieback with typical symptoms. Although multiple plant pathogenic Diaporthe species have been identified in China, little is known about economically and ecologically valuable species [27]. In addition, D. phaseolorum is an endophyte found in mangrove forests [28]. The closely related taxa D. endophytica, D. schini, and D. terebinthifolii were originally described as endophytes from Schinus spp. in Brazil [6]. Additionally, Diaporthe spp. are known to be infectious, particularly in people with weakened immune systems; however, they are rare diseases of major medical importance [29]. D. endophytica was found to be an endophytic from the medicinal plants S. terebinthifolius and Maytenus ilicifolia in Brazil [6], however, D. endophytica can control the pathogen Phyllosticta citricarpa that is responsible for citrus black spot in citrus plants [30]. However, Diaporthe species associated with plant and forest tree diseases have rarely been studied in Korea. Therefore, specimens must be collected from a wide variety of habitats and hosts to fully understand the taxonomy of Diaporthe species. KNU-JJ-1809 was isolated from soil and identified as D. endophytica which has not been previously reported in Korea.
Taxonomy
Faurelina indica Arx, Mukerji & N. Singh, Sydowia 34:39 (1981) (Fig. 3).
Ecology and distribution: Very few members of this genus have been reported to date. Some species were identified in the fruit of herbivore dung collected from the Caatinga biome in the semi-arid region of Brazil (F. fimigena and F. hispanica). The type species F. indica was isolated from the dung of cows and goats (India). Some members of this genus have been identified in Japan and France. The present studied strain KNU-JJ-1830 was collected from soil in Korea.
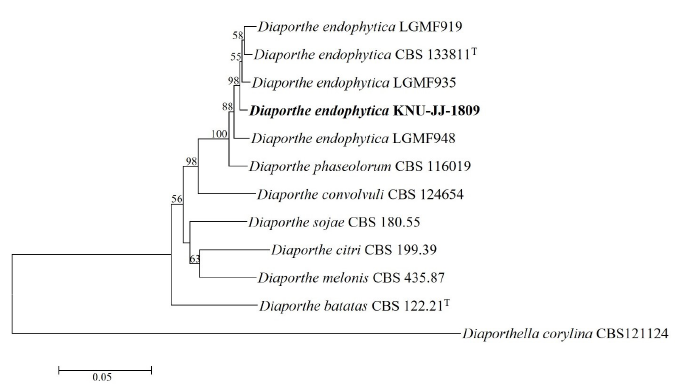
Fig. 2. Neighbor-joining phylogenetic tree of the strain KNU-JJ-1809 based on the combined sequences (ITS+TUB2+CAL+TEF1α ), showing the relationships between Diaporthe endophytica with the closest species of Diaporthe. Diaporthella corylina CBS 121124 was used as an outgroup. Numbers above the branches represent the bootstrap values obtained for 1,000 replicates (values smaller than 50% are not shown). The isolated strain of this study is indicated in bold. Bar, 0.05 substitutions per nucleotide position. ITS, internal transcribed spacer of nuclear ribosomal DNA; TUB2, β-tubulin gene; CAL, calmodulin gene; TEF1α , translation elongation factor 1-alpha gene.
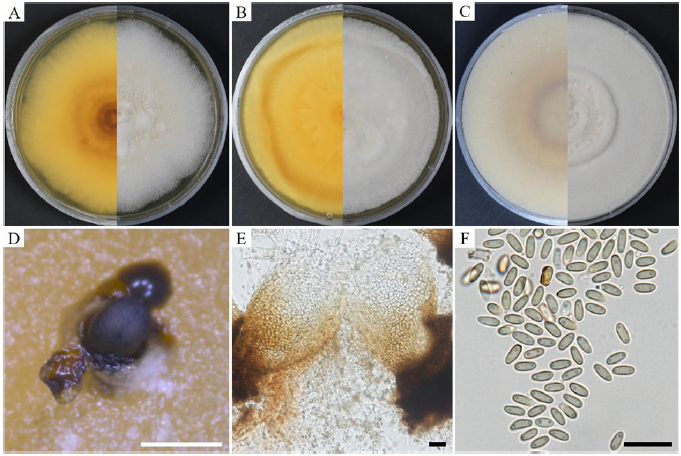
Cultural and morphological characteristics: To study the cultural and morphological characteristics, the strain was cultured on PDA, MEA, and OA media (Table 3). The colonies were white to pale yellow, reaching 80-85 mm in diameter on PDA after 21 days at 25℃, and reverse yellowish to dark yellow toward the center (Fig. 3A). On MEA, the colonies were white to pale yellow, reverse yellow, yellow patches toward the edge, and irregular patches near the center, covering 85-90 mm in diameter after 21 days at 25℃ (Fig. 3B). On OA, the colonies were white, smooth, with tufts of irregular aerial mycelia toward the center, reverse light brown to brown toward the center, and covered the Petri dishes after 21 days at 25℃ (Fig. 3C). Conidiomata were immersed at the base, dark black, hemispherical or pustulate, rounded above, smooth, and 200-350 μm in size (Fig. 3D). The vegetative hyphae were hyaline or light brown, thick-walled, branched, and septate. The pycnidial walls released mature yellowish conidia (Fig. 3E). Conidia were hyaline to light green, thin, clavate, round, truncate base, with guttules in both ends, and with a diameter (n=100) of 2.5-5.2×1.7-3.8 μm (average, 4.5×2.0 μm) (Fig. 3F). Based on previous studies, KNU-JJ-1830 was found to be similar to F. indica based on its cultural and morphological characteristics (Table 3).
Molecular phylogeny of KNU-JJ-1830
Nucleotide sequences of the ITS (547 bp) and 28S ribosomal DNA (883 bp) regions were obtained after sequence analysis. The 28S rDNA showed 98.98% and 99.21% identity with F. indica strains CBS 126.78 and CBS 301.78, respectively. The 28S ribosomal DNA dataset obtained using the NJ phylogenetic algorithm showed that KNU-JJ-1830 and the reference strains of F. indica (CBS 126.78 and CBS 301.78) produced clusters with a high bootstrap value (100%) (Fig. 4). Thus, KNU-JJ-1830 was identified as F. indica.
The type species of F. indica and F. elongata (previously known as Leuconeurospora elongata) were previously identified in goat and cow dung collected in Nainital, Delhi, India [12]. Although one of these species was L. elongata, it was later transferred to the genus Faurelina [10]. F. elongata was obtained from Kagoshima (southwestern Kyushu, Japan). Moreover, the fruiting of two species, F. fimigena and F. hispanica, was recorded in herbivore dung collected from the Caatinga biome in the semi-arid region of Brazil [31]. However, a lack of knowledge about fungal diversity, particularly coprophilous fungi, is an important factor that complicates conservation strategies. A focused study of herbivore dung mycobiota can improve our knowledge of the biogeography and diversity of numerous unreported species. Few species have been identified in the genus Faurelina and have been recorded worldwide. This is the first recorded genus and the first record of F. indica in Korea.
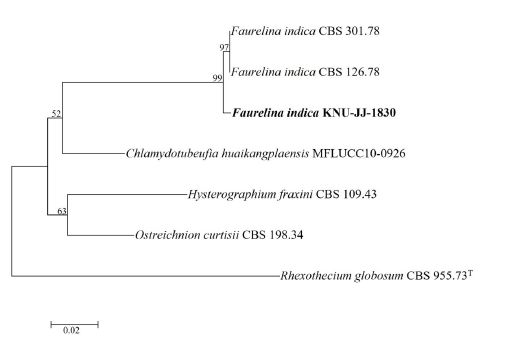
Fig. 4. Neighbor-joining phylogenetic tree of the strain KNU-JJ-1830 based on sequences of 28S ribosomal DNA to reveal the phylogenetic position of Faurelina indica . Rhexothecium globosum CBS 955.73T was used as an outgroup. Numbers above the branches represent the bootstrap values obtained for 1,000 replicates. The isolated strain of this study is indicated in bold. Bar, 0.02 substitutions per nucleotide position.
Taxonomy
Trichoderma ivoriense Samuels, Mycological Progress 11:237 (2011) (Fig. 5).
Ecology and distribution: Members of this genus have been observed on several hosts, such as mushroom compost (Canada, Ireland, England, USA), soil in kiwifruit orchards (New Zealand), forest soil (USA), Quercus infected with Lentinus edodes (Japan), botanical garden soil (England), maize field soil (China, Colombia), Pleurotus spawn (Netherlands), soil (Japan, Cameroon), water-damaged buildings (Denmark), and vegetable greenhouse soil (China). The present studied strain KNU-4-KH1 was isolated from soil in Korea.
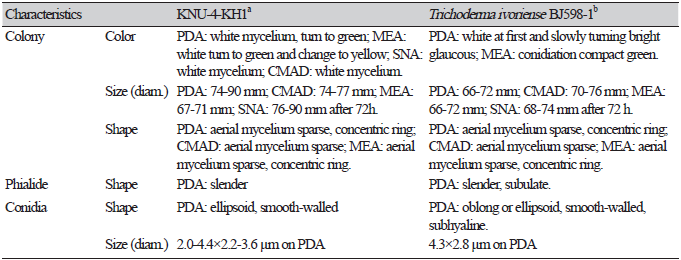
Cultural and morphological characteristics: KNU-4-KH1 was cultured at 25℃ for 72 h in PDA, CMAD, MEA, or SNA media (Table 4). Consequently, mycelial growth was 74.4-77.7, 74.0-90.0, 67.5-71.5, and 82.0-90.0 mm, respectively. The mycelia initially appeared white, turned pale green, and later became green on PDA (Fig. 5A). The hyphae were sparse and hyaline to greenish with two concentric circles, which became thicker. Several green and white pustules were observed in the medium. CMAD did not change color to white hyphae, even after one month (Fig. 5B). On MEA, the mycelia turned from white to very pale green over time, gradually turned yellow, and had two concentric circles, similar to that on PDA (Fig. 5C). On SNA medium, the hyphae grew close to the medium surface, and green pustules were observed around the incised area (Fig. 5D). Conidiophores in the pustules had long, sterile hairs that were light brown to blackish in color and short, fertile branches growing from their bases (Figs. 5E and F). Fertile branches growing from the base of the hair normally had a single basal cell and produced terminal phialides, whereas branches farther from the tip typically did not have a single basal cell and did not produce terminal phialides (Figs. 5G and H). Numerous conidia were observed on the PDA. Conidia were oblong or ellipsoidal, smooth-walled, greenish, and with a diameter (n=50) of 2.0-4.4×2.2-3.6 μm (Fig. 5I). Strain KNU-4-KH1 was confirmed to be a previously identified T. ivoriense strain based on its cultural and morphological characteristics (Table 4).
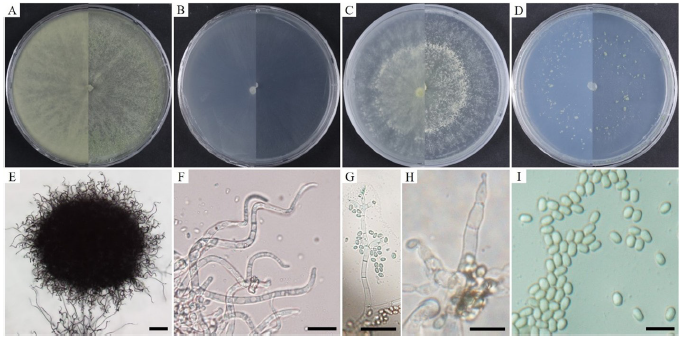
Fig. 5. Cultural and morphological characteristics of Trichoderma ivoriense KNU-4-KH1. Colony on potato dextrose agar (A); corn meal agar (B); malt extract agar (C); and synthetic nutrient-poor agar (D); Hairs on conidial pustules (E, F); fertile branch with phialides (G, H); and conidia (I). Scale bars: E=100 μm; F-I=10 μm.
Molecular phylogeny of KNU-4-KH1
After the sequence analysis of KNU-4-KH1, 549 bp from the ITS region, 586 bp from TEF1α, and 459 bp from CAL were obtained. BLAST analysis of the ITS sequences indicated a maximum identity of 100% with the strains of Trichoderma ivoriense (CBS 125734T, BJ598-1). TEF1α showed maximum 98.7% and 99.3% similarities with the CBS 125734T and BJ598-1 strains of T. ivoriense, respectively. In addition, the CAL gene showed the maximum 99.6% similarity to T. ivoriense CBS 125734T. Phylogenetic analysis of the combined dataset of ITS, TEF1α, and CAL revealed that the strain from this study and the reference strain of T. ivoriense CBS 125734T clustered together with a high bootstrap value (100%) in the NJ phylogenetic tree (Fig. 6). Thus, KNU-4-KH1 was identified as T. ivoriense.
The genus Trichoderma is found in various parts of the world on various substrates. The physicochemical mechanisms used by Trichoderma spp. to obtain nutrients allow them to thrive in a wide variety of substrates and environmental settings [12]. There are multiple species in the genus Trichoderma, making it hyperdiverse and with a global reach, spreading over many different substrates and locations [32]. This genus has proven to be useful in agriculture (biocontrol of plant pathogens), industry (prevention of pollution), and environmental protection [33,34]. Many researchers worldwide are interested in this group owing to its diversity and the resources it continues to explore. However, some species are responsible for the spread of green mold illness during mushroom farming [35] and even serve as opportunistic human diseases [36]. Although the genus Trichoderma serves as one of the best bioagents effective against a wide range of pathogens, the exact role of T. ivoriense as a biological control agent remains unknown. Therefore, the biological control activity of the isolated strain T. ivoriense requires further examination. To the best of our knowledge, this is the first report of T. ivoriense in Korea.
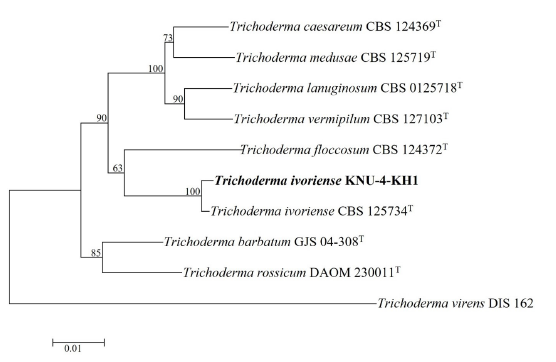
Fig. 6. Neighbor-joining phylogenetic tree of the strain KNU-4-KH1 based on the combined sequences (ITS+TEF1α+CAL), showing the relationships between Trichoderma ivoriense with the closest species of Trichoderma . Trichoderma virens DIS 162 was used as an outgroup. Numbers above the branches represent the bootstrap values obtained for 1,000 replicates. The isolated strain of this study is indicated in bold. Bar, 0.01 substitutions per nucleotide position. ITS, internal transcribed spacer of nuclear ribosomal DNA; TEF1α , translation elongation factor 1-alpha gene; CAL, calmodulin gene.