Basil (Ocimum basilicum L.) is an annual or subshrub belonging to the family Lamiaceae, and its native range is tropical and subtropical Asia to North Australia [1]. This plant grows primarily in the seasonally dry tropical biomes and is used worldwide as a culinary herb or medicine [2]. In Korea, this plant is cultivated as an herb or vegetable in vinyl greenhouses.
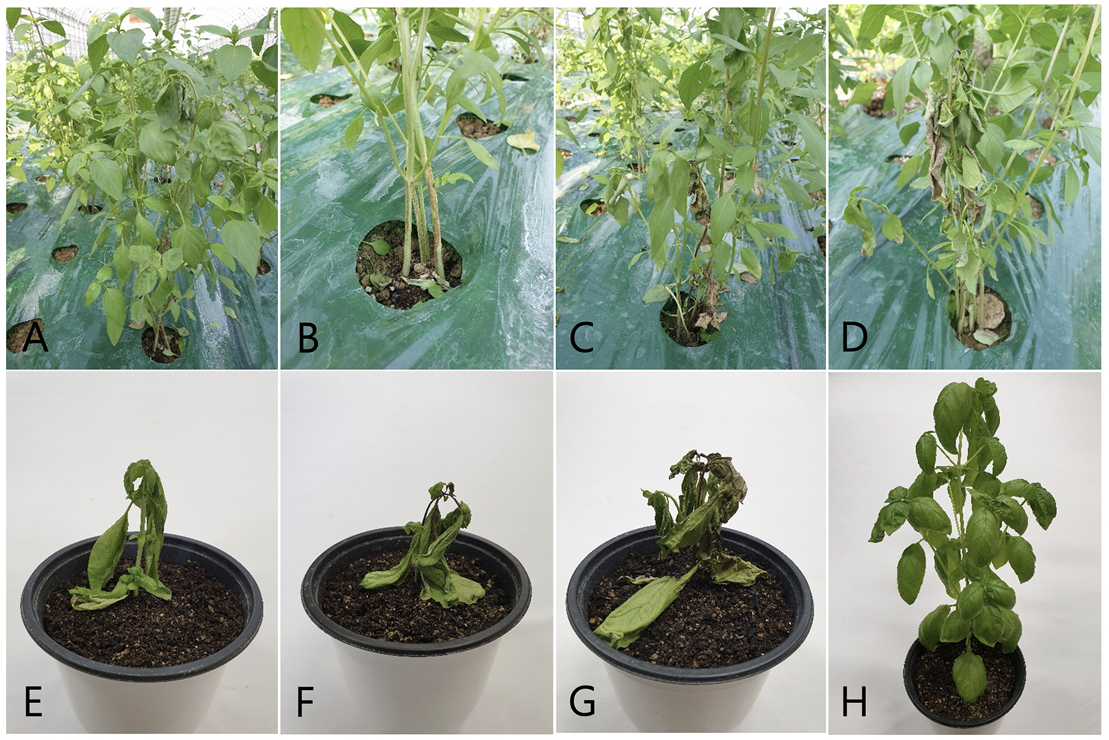
We observed wilt symptoms in basil plants grown in a vinyl greenhouse located in Gokseong, Korea, during crop disease surveys conducted in August 2022. In the early stages of the disease, the symptoms appeared as slight wilting of the leaves and apices and brown to dark brown longitudinal streaks on the stems at or above the soil line (Fig. 1A and B). Severely diseased plants wilted wholly and were blighted with dark discoloration (Fig. 1C and D). Three sites were observed in the vinyl greenhouse, and 50 plants at each site were investigated for the disease incidence. The incidence of the disease in the plants ranged from 5 to 20%.
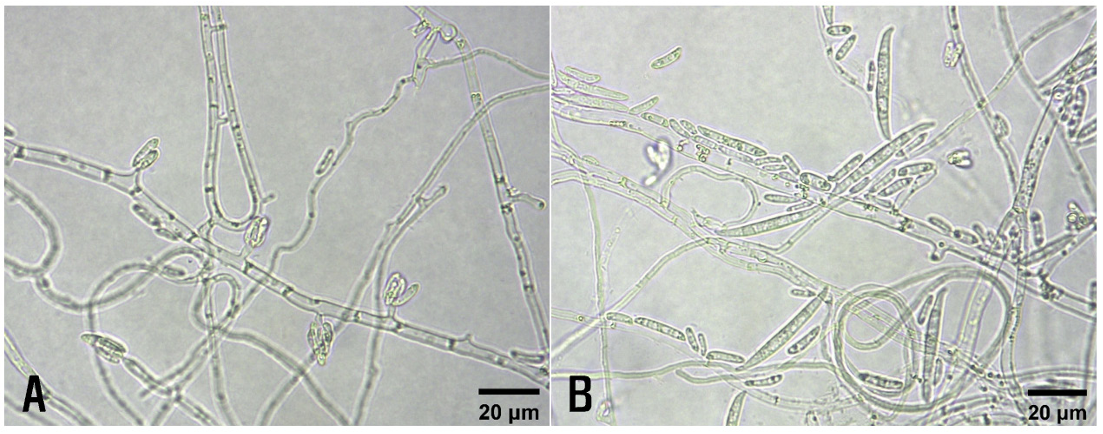
Basil stems with wilt symptoms were collected from the investigated vinyl greenhouse, and fungi were isolated from the diseased stems. The 3-5 mm-long lesion pieces cut from the stems were plated on 2% water agar after surface sterilization with 1% sodium hypochlorite solution for 1 min. The fungal mycelia growing from the lesion pieces were transferred to potato dextrose agar (PDA) slants after incubating the plates at 25℃ for 2-3 days. Six isolates of Fusarium sp. were obtained from the lesion pieces, and their morphological characteristics were examined under a light microscope (Eclipse Ci-L; Nikon, Tokyo, Japan). Microconidia and macroconidia were produced on short monophialides (Fig. 2A and B). Microconidia were ellipsoidal to cylindrical, straight to curved, 0-1 septate, and measured 6.0-17.0×1.5-4.0 μm (av. 9.7×2.5 μm). Macroconidia were falcate, foot-shaped at both ends, 2-5 septate, and measured 20.0-45.0 ×2.5-4.8 μm (av. 30.2×3.8 μm). The size of the conidiophores was 2.7-19.0×1.0-4.0 μm (av. 6.2× 2.2 μm). The morphological characteristics of these isolates were similar to those of Fusarium oxysporum Schltdl. as previously described [3-5]. Among the isolates, two (OBFU-01 and OBFU-06) were used for phylogenetic analysis and pathogenicity test.
Fig. 2
Morphological features of Fusarium oxysporum isolates from diseased basil plants. (A) Microconidia, monophialides, and hyphae of the isolate. (B) Microconidia, macroconidia, monophialides, and hyphae of the isolate.
Phylogenetic analysis was conducted to confirm the identity of F. oxysporum isolates based on their morphological characteristics. Genomic DNA was obtained from the isolates using a previously described protocol [6], with slight modifications. In polymerase chain reaction (PCR) experiments, partial translation elongation factor 1-α (TEF1) and RNA polymerase II second largest subunit (RPB2) gene regions were amplified using primer sets of EF-1 and EF-2 for TEF1 [7], and 5F2-7cR and 7cF-11aR for RPB2 [8]. PCR products were prepared using the DNA Free-Multiplex Master Mix (Cellsafe, Yongin, Korea) and amplified as described in the previous studies [8,9]. The PCR products were purified using the Universal DNA Purification Kit (Tiangen, Beijing, China). PCR products were sequenced at Bionics (Seoul, Korea) using the same primers. Sequences were adjusted using SeqMan II (DNASTAR Inc., Madison, WI, USA), whenever necessary.
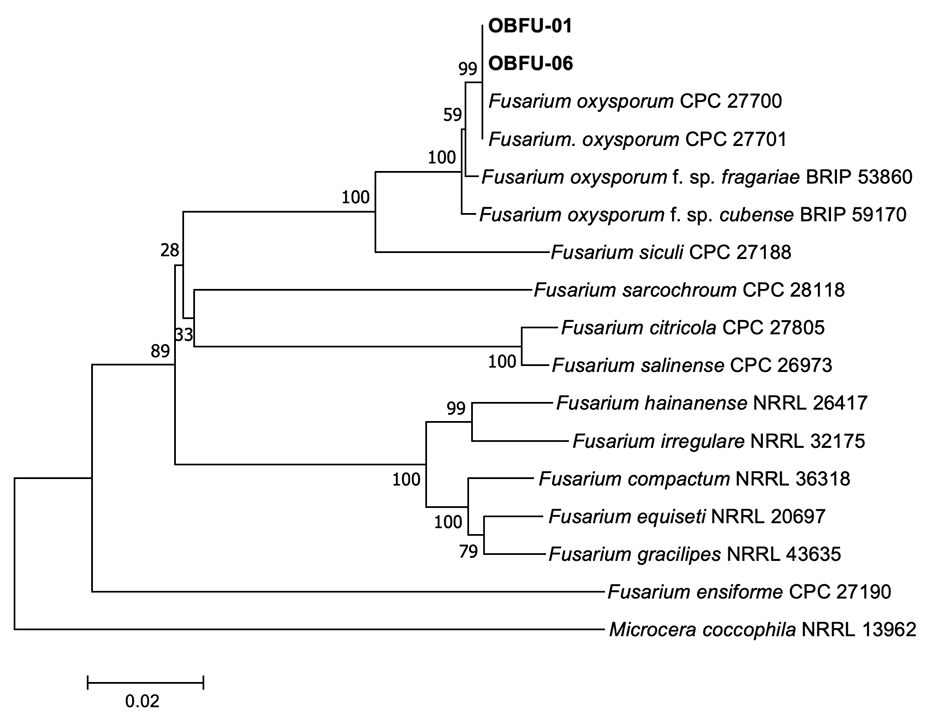
The sequences of the isolates (OBFU-01 and OBFU-06) obtained from basil plants and relevant sequences of Fusarium spp. from Genbank were aligned using MUSCLE [10]. Alignments were then improved using MEGA version 7 software [11], if necessary. Concatenated alignments were used for neighbor-joining analysis with the maximum composite likelihood model conducting 1,000 bootstrap replicates using the MEGA version 7 software [11]. Microcera coccophila (NRRL 13962) was selected as the outgroup taxon. Bootstrap values were shown at the nodes. Phylogenetic analysis based on sequence alignments of TEF1 genes revealed that the isolates clustered with two strains (CPC 27700 and CPC 27701) of F. oxysporum and two strains (NRRL 38318 and FOB08) of F. oxysporum f. sp. basilici Tamietti & Matta (Fig. 3). Phylogenetic analysis based on concatenated sequence alignments of TEF1 and RPB2 genes revealed that the isolates clustered with two strains (CPC 27700 and CPC 27701) of F. oxysporum (Fig. 4). The sequence data of TEF1 and RPB2 genes obtained from the isolates were deposited in NCBI Genbank under the accession numbers OR866375–OR866376 and OR866377–OR866378, respectively.
Two isolates (OBFU-01 and OBFU-06) of F. oxysporum were tested for pathogenicity in basil plants by
Fig. 3
Phylogenetic tree based on a concatenated alignment of partial translation elongation factor 1-α sequence data of the isolates (OBFU-01 and OBFU-06) from diseased basil plants and reference strains of Fusarium species. The phylogenetic tree was generated using the neighborjoining method with the maximum composite likelihood model. Bootstrap support values are indicated at the nodes. The scale bar represents the number of nucleotide substitutions per site.
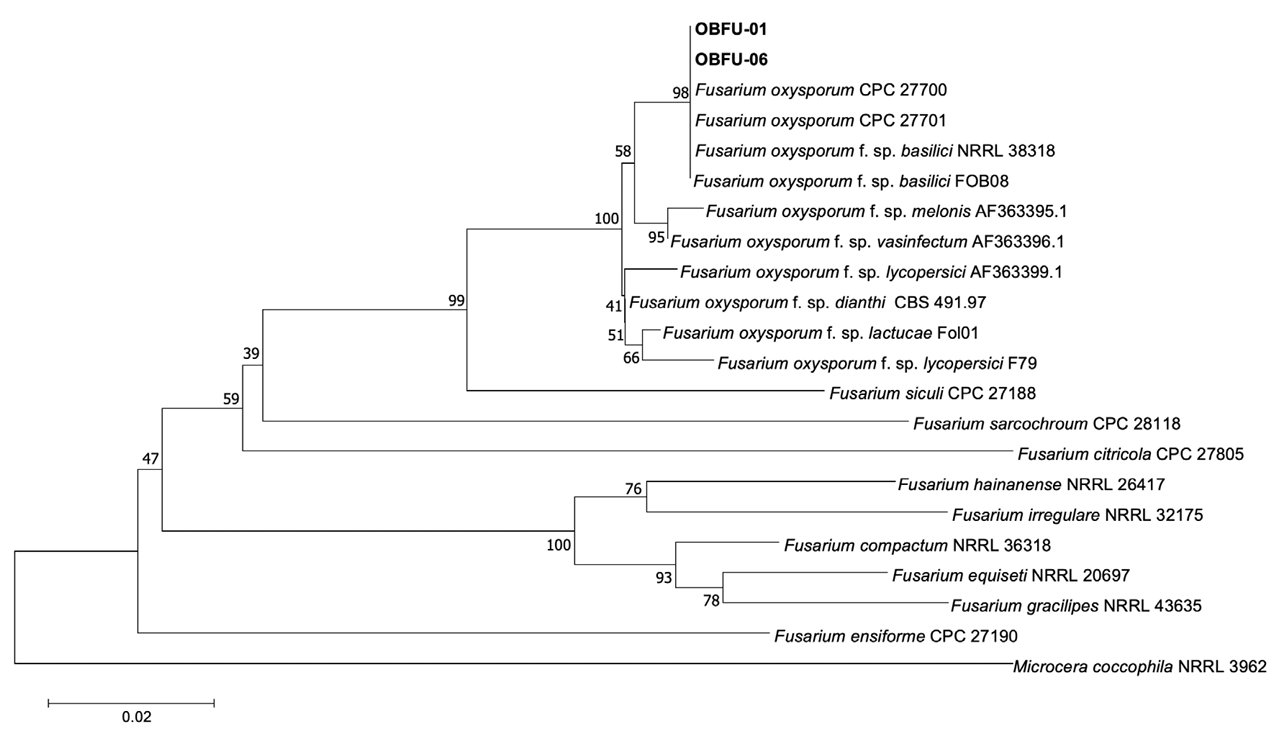
Fig. 4
Phylogenetic tree based on a concatenated alignment of partial translation elongation factor 1-α and RNA polymerase II second largest subunit sequence data of the isolates (OBFU-01 and OBFU-06) from diseased basil plants and reference strains of Fusarium species. The phylogenetic tree was generated using the neighbor-joining method with the maximum composite likelihood model. Bootstrap support values are indicated at the nodes. The scale bar represents the number of nucleotide substitutions per site.
Many species complexes have been reported in the genus Fusarium [12], and F. oxysporum has been reported to be a species complex [13-15]. The epitype of F. oxysporum was established using multilocus phylogenetic inference and subtle morphological differences [5]. In this study, Fusarium sp. isolates from diseased basil plants were identified as F. oxysporum based on the morphological characteristics and phylogenetic analyses.
F. oxysporum causes wilt in many plants [3,16]. It has been reported that formae speciales exist in F. oxysporum species complex [17]. F. oxysporum f. sp. basilici was named as a pathogen of Fusarium wilt of basil [18] and has also been reported to cause Fusarium wilt and crown rot in sweet basil [19]. However, phylogenetic analysis based on sequence alignments of TEF1 genes revealed that the two strains of F. oxysporum f. sp. basilici clustered with the two strains of F. oxysporum and our isolates, suggesting that forma specialis basilici of F. oxysporum should be reconsidered. We could not obtain the sequence data for RPB2 genes of F. oxysporum f. sp. basilici from Genbank. Therefore, further studies are needed to clarify the forma specialis of F. oxysporum that causes Fusarium wilt in basil. F. oxysporum has been reported to cause Fusarium wilt in many plants in Korea [20]. However, there are no reports on the occurrence of Fusarium wilt in basil caused by the fungus in Korea. This is the first report of F. oxysporum causing Fusarium wilt in basil in this region.