Rice is the second-most important cereal food crop after wheat, with Asia accounting for 90% of rice produced worldwide [1]. In Korea, rice comprises approximately 70% of dietary cereal and grain intake [2]. Rice production faces numerous challenges because of the frequent emergence of pests and diseases. Rice sheath rot disease, an emerging rice panicle disease capable of causing yield losses of 20-85%, has been reported in almost all rice-growing areas worldwide [2]. The infected leaf sheath typically rots and turns grayish-brown or reddish-brown, depending on the rice cultivar, leading to unmerged and empty panicles. The major causative agent was identified as Acrocylindrium oryzae, which was later named as Sarocladium oryzae. However, other studies showed that Fusarium spp. belonging to the F. fujikuroi species complex and the bacterial pathogen Pseudomonas fuscovaginae can also cause sheath rot in rice. These organisms produce a range of phytotoxins that induce necrosis in host plants. Pathogens in the genus Sarocladium (S. oryzae, S. attenuatum, S. sparsum, S. strictum) are mostly found in lowland environments, where hot and humid weather favors their establishment [3]. Under these conditions, the pathogen can survive in infected seeds, plant residues, soil, water, and weeds. The pathogen can infect rice at various growth stages by entering the plant stomata or wounds, with signs and symptoms of infection appearing during or after the booting stage. Insect and mite damage or weakening of the plant by other pathogens facilitates entry of Sarocladium sp. into the plant [3]. Secondary infections may be windborne when the pathogen enters the plant through injured tissues. Sarocladium spp. mainly use rice as a host but have also been reported to be the causal agents of bamboo blight in bamboo plants, grasses, and sedges in Bangladesh and India [3]. In China, S. strictum is associated with brown spots on apple [4]. In Korea, S. oryzae was reported to be involved in rice ear blight; however, whether Sarocladium sp. is involved in sheath rot disease is unknown [5]. Therefore, studies are needed to understand the impact of yield loss caused by sheath rot disease in rice and to incorporate an integrated disease management approaches in Korea.
Hence, this study was conducted to determine whether Sarocladium spp. is present in Korea as well as the pathogenicity and cultural characteristics of the pathogens through morphological and molecular studies. In September 2023, rice showing sheath rot was observed in rice fields at the National Institute of Agricultural Sciences, Korea (35°82'88.0"N, 127°04'44.0"E). The rice sheath had lesions with pale gray and brown centers and dark brown margins (Fig. 1A and B). To isolate the fungus, the panicles and excised lesions were disinfected with 1% sodium hypochlorite for 3 min and placed on water agar; the plates were incubated at room temperature. Sarocladium strictum produced mycelia and conidial chains on surfacesterilized rice sheath lesions (Fig. 1C). At 10 days post-inoculation, pure cultures were obtained by placing small agar blocks containing single hyphal tips onto potato dextrose agar (PDA) plates. Both acquired strains, FD00214 and FD00215, were preserved on PDA slants at the Plant disease Laboratory (National Institute of Agricultural Sciences, Wanju, Korea). Agar blocks were cut from the mycelial fronts of the PDA slants, placed at the center of PDA plates, and incubated at 25℃ in the dark. Mycelial growth was measured at 14 days post-inoculation, followed by observation under a light microscope. Measurements and photographs were taken using slide preparations with water as the mounting medium. Morphological characteristics were examined at 14 days post-incubation at 25℃ in the dark. The colonies were moist to smooth, pale orange to pink in color, and 40 mm in diameter (Fig. 2A and B). The phialides hyaline were slender, smooth walled and measured 10-32 (21.7)×2-4 (2.7) μm in diameter (Fig. 2C). Conidia were one-celled with a slimy head and measured 3-6 (4.5)×2-4 (2.4) μm in diameter (Fig. 2D). The morphological and culture characteristics of the isolates were similar to those previously reported for Sarocladium species [6,7]. The isolates FD00214 and FD00215 were cultured in 20 mL potato dextrose broth for 10 days, and DNA from representative isolates was extracted using a Maxwell® RSC PureFood GMO and Authentication Kit (Promega, Madison, WI, USA). The internal transcribed spacer (ITS) and actin gene (ACT) regions were amplified using primer pairs ITS1/ITS4 and Act1/Act4, respectively [8,9]. The PCR program was performed as follows: denaturation at 94℃ for 2 min; followed by amplification for 35 cycles of 94℃ for 30 s, 55℃ for 30 s, and 53℃ for 30 s; and 72℃ for 60 s. Final extension was performed for 10 min at 72℃. The 25 µL reaction mixture contained 1 µL each F/R primer (10 pmol/ µL), 2.5 µL 10x EasyTaq (buffer), 1 µL dNTPs, 0.2 µL Taq DNA polymerase, 1 µL template DNA (33 ng/µL), and 19.3 µL ddH2 O. The amplified products were purified using agarose gel electrophoresis and then sequenced. The sequences of the ITS and ACT genes were compared with those in GenBank using BLASTn (https://blast.ncbi.nlm.nih.gov/Blast.cgi). The resulting sequences were deposited in GenBank under accession nos. LC784015–LC784018. A BLASTn search of the ITS and ACT DNA sequences showed 99.81% and 98.32% identity to S. strictum isolate CBS 346.70, respectively. Reference sequences of the ITS and ACT genes were used to generate a phylogenetic tree [10]. Sequences were assembled using the MUSCLE program (Edgar 2004) and edited manually using MEGA X [11]. The alignment consisted of seven ITS and ACT gene sequences of Sarocladium spp., including the reference sequences. A phylogenetic tree was constructed using the maximum likelihood method and implemented in Mega XI software with 1,000 bootstrap replications [12]. The reference sequence for Acremonium curvulum was used as an outgroup in the phylogenetic tree. The amplified and sequenced PCR products of Sarocladium sp. isolates were approximately 600 and 900 bp long for the ITS and ACT gene regions, respectively. The phylogeny inferred using the sequence data obtained in this study, together with sequence data from Sarocladium species previously described by Abdollahi and Fotouhifar (2016), placed our isolates together with S. strictum (syn. Acremonium strictum) isolates (Fig. 3). The results of phylogenetic analysis were consistent with the description of S. strictum. A pathogenicity test was conducted using the attached tiller assay inoculation technique [13]. Briefly, a seven-day old culture of S. strictum culture on PDA was used. The mycelial mat on the PDA medium was cut into approximately equal sizes using a sterile blade and used for inoculation. Three tillers (per plant) at the booting stage, grown in pots inside a screenhouse, were inoculated with mycelial discs, covered with sterile moistened absorbent cotton, wrapped with paraffin film, and wrapped with PDA disks without mycelia. The plants were maintained at high humidity. Symptoms were observed daily to verify that the initial symptoms of S. strictum were re-isolated using PDA plates for the inoculated isolates. Symptoms included lesions appearing as grayish-brown and reddish-brown, along with unmerged and empty panicles only on the inoculated leaf sheath at 14 days post-inoculation, whereas control samples remained asymptomatic (Fig. 4). The S. strictum isolates were re-isolated from the infected tissues of the inoculated leaves, thus fulfilling Koch's postulates. Our findings are similar to those of recent studies of the interactions between maize and S. strictum isolates from Cameroon, which led to detection of a species with pathogenic properties [14]. Sarocladium strictum was also found to cause wilting and death of strawberry plants [15]. The isolates FD00214 and FD00215 were identified as S. strictum based on their morphology and molecular data. This species is frequently isolated from soil and plant rhizospheres as a hyperparasitic fungus. This fungus was also recorded as Acremonium strictum from Heterodera schachtii and some plant substrates, such as Vitis sylvestris and Zea mays in Iran [16]. To the best of our knowledge, this is the first report of sheath rot of rice caused by S. strictum in South Korea.
Fig. 1
Sheath rot caused by Sarocladium strictum on Rice (Oryza sativa). A, symptoms on the leave sheath having pale grey/brown centers with dark brown margins along the sheath disrupting panicle emergence under in field condition. B, rotting of rice panicles inoculated with S. strictum at booting stage (White arrows=symptom in rice). C, Conidia- grouped in slimy heads. Scale bar C=10 µm.
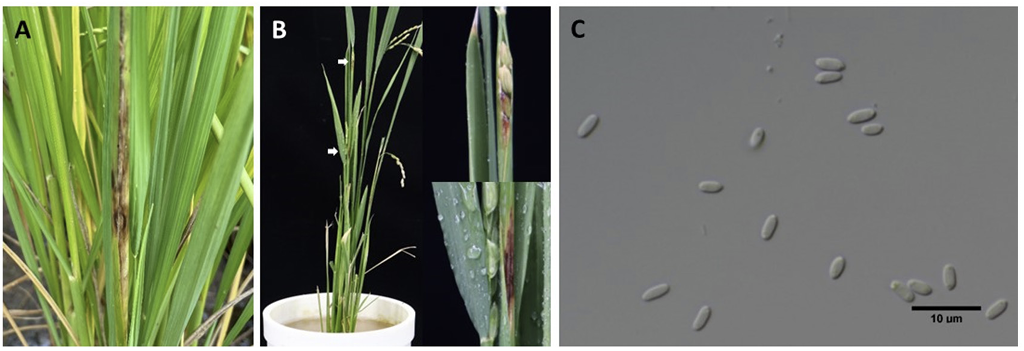
Fig. 2
Sarocladium strictum colony on potato dextrose agar (PDA) incubated at 25℃ in the dark for fourteen days. A, observed colony. B, reverse side of the colony. C, slender smooth walled phialide hyaline. D, 1–celled, cylindrical conidia- grouped in slimy heads. Scale bar C=10 µm; D=10 µm.
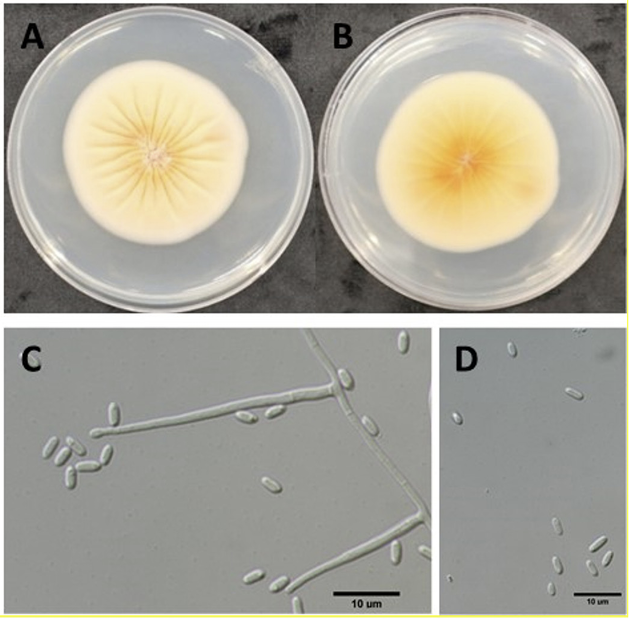
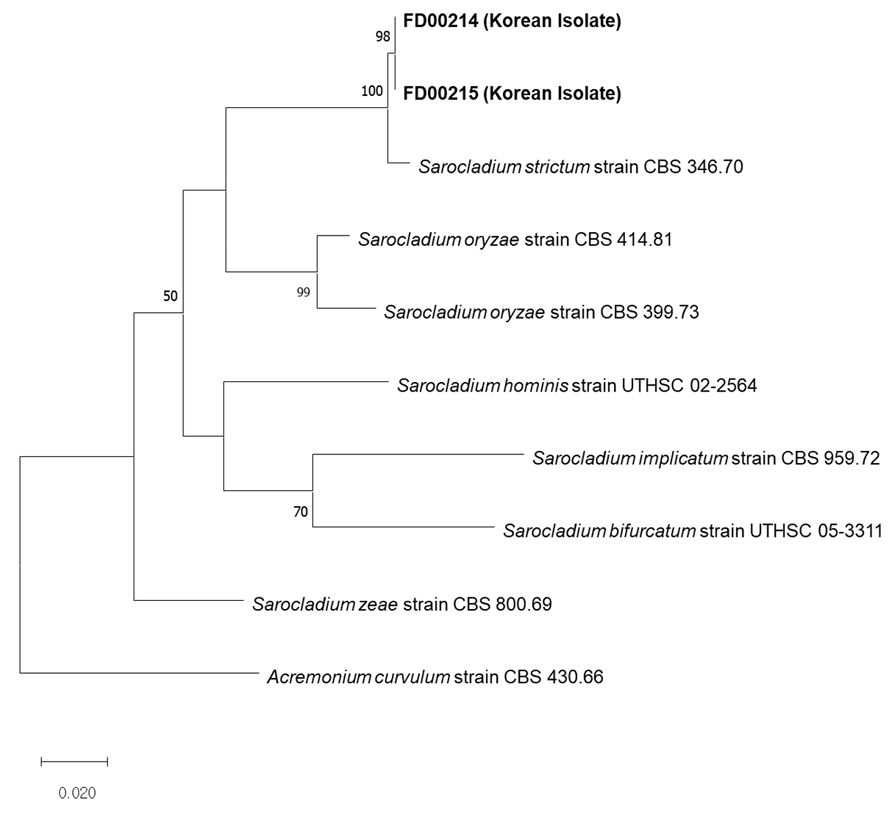
Fig. 3
Maximum likelihood phylogenetic analyses based internal transcribed spacer (ITS), and actin gene (ACT) sequence. The studied Sarocladium isolates are shown in bold. The isolates formed a monophyletic group with S. strictum isolate CBS 346.70. Acremonium curvulum strain CBS 430.66 was used as an outgroup. Bootstrap value under 50% is not shown.