INTRODUCTION
Yeasts are unicellular fungi that reproduce by budding or fission and are not a taxonomic classification. They possess specific morphological and physiological traits that distinguish them from other fungi [1,2]. Yeast has a rich history in the production of traditional fermented foods (particularly alcohol) that was utilized approximately 9,000 years ago. Recent studies show that yeasts are a versatile group of organisms with the potential for use in various industries. They can produce a range of bioactive substances, bioethanol, pigments, and enzymes, and serve as hosts in molecular biology [3,4].
In the past, yeasts found in Korea was primarily isolated and identified from fermented foods such as traditional soybean paste or meju. However, there is a recent increase in the isolation and characterization of yeasts from natural environments (including flowers and fruits) for application in numerous industries [5-8]. These studies indicate that wild yeasts in natural environments exhibit species diversity based on their source of isolation, which varies according to the climatic characteristics of the region. Furthermore, the same source of isolation can yield different yeast communities, depending on the collection location [3].
In this study, we isolated and identified wild yeasts from flowers, trees, and fruits collected in the natural environment in Korea, including various botanical gardens and parks in Yongin, Suwon, Daegu, Hongseong, Jeju, and Osan. Finally, we have selected 12 unrecorded species of yeasts that are not included in the National Species List of Korea (NSLK), namely Sakaguchia oryzae, Cystobasidium raffinophilum, Meira argovae, Kazachstania humilis, Meyerozyma smithsonii, Anthracocystis trispicatae, Naganishia brisbanensis, Tremella yokohamensis, Kwoniella shandongensis, Kwoniella newhampshirensis, Aureobasidium proteae, and Rhodotorula dairenensis. The microbiological characteristics of these yeast strains are described in detail.
MATERIALS AND METHODS
Species and strain selection
Four hundred and fifty yeast strains were isolated from various regions and materials such as Zelkova, Quercus oak, Camellia, and tangerine trees in Korea. Among them, twelve selected yeast strains not recorded in the NSLK were selected for listing. Samples were collected in sterilized tubes from different locations, crushed with a mortar and pestle and soaked overnight in 10 mL saline solution (pH 3.3) to obtain a suspension. The suspension was spread on yeast extract peptone dextrose (YPD) agar containing ampicillin (0.1 mL/100 mL), dichloran rose bengal chloramphenicol (DRBC) agar, and dichloran glycerol (DG18) agar, as previously described [9]. Yeast colonies were grown for 48 h at 25℃, then picked and isolated as a single colony for identification [10].
DNA isolation, amplification, and phylogenetic analysis
The isolated yeast strains were identified by analyzing the sequence of the internal transcribed spacer (ITS) region with biological relationships registered in the National Center for Biotechnology Information (NCBI) database. Polymerase chain reaction (PCR) was performed to amplify the ITS region using the ITS1 (TCCGTAGGTGAACCTGCGG) and ITS-4 (TCCTCCGCTTATTGATATGC) primers. The reaction was performed in the following steps: pre-denaturation at 95℃ for 5 min, 30 cycles of denaturation at 95℃ for 30 s, annealing at 55℃ for 30 s, and extension at 72℃ for 30 s. Finally, a post-extension step was performed at 72℃ for 10 min. The ITS region sequences of the isolates were deposited in GenBank (https:// www.ncbi.nlm.nih.gov/genbank/).
Phylogenetic analysis was performed for taxonomic classification using the ITS region nucleotide sequences of the type strains of species within the genus or family from the NCBI database. A phylogenetic tree was constructed using MEGA X software through the neighbor-joining method, which relies on the Tamura-Nei model. Bootstrap analysis was performed 1,000 times to ensure accuracy [11].
Morphology
Cell morphology was observed by culturing cells on yeast malt (YM) agar plates for 3 d at 25℃ and observing them using a light microscope (DN-10A, Samwon, Goyang, Korea). Colony formation was observed by inoculating the single colony and growing on YM agar plate for 7 d at 25℃. The formation of hyphae and pseudohyphae was observed using Dalmau plates on cornmeal agar for 14 d at 25℃ [12].
RESULTS AND DISCUSSION
The origin of 12 undocumented yeast strains identified in this study are shown in Table 1. These 12 strains consisted of three Ascomycota and nine Basidomycota yeasts. These strains were identified by comparison with the ITS region sequence of the type strain. Consequently, the yeast strain Naganishia brisbanensis KHU20220630-01 (Genbank: OR791288) obtained from Liriope muscari produced a single clade with the N. brisbanensis holotype strain BRIP 28244 (Fig. 1A). Similarly, the yeast strain Sakaguchia oryzae KHU20220630-02 (Genbank: OR791290) obtained from Pteridium aquilinum var. latiusculum formed a single clade with the type strain CBS 9745 of S. oryzae (Fig. 2A). The yeast strain Cystobasidium raffinophilum KHU20220630-03 (GenBank: OR791282) from Camellia japonica formed a single clade with the holotype strain CGMCC 2.3822 of C. raffinophilum (Fig. 3A), and the yeast strain Meira argovae KHU20220630-04 (GenBank: OR791287) from Olea europaea formed a single clade with the type strain CBS 110053 of M. argovae (Fig. 4A). Kazachstania humilis KHU20220630-05 (GenBank: OR791284) obtained from C. reticulata×C. sinensis formed a single clade with K. humilis type strain CBS 5658 (Fig. 5A). Meyerozyma smithsonii KHU20220630-06 (GenBank: OR791289) from Citrus reticulata confirmed the presence of two sequences within the same species in the database and produced a single clade using M. smithsonii strain H1 instead of the type strain (Fig. 6A). Anthracocystis trispicatae KHU20220630-07 (GenBank: OR791280), obtained from Bambusoideae formed a single clade with isotype strain BRIP 47730 of A. trispicatae (Fig. 7A). Tremella yokohamensis KHU2023032801 (GenBank: OR791291) from Quercus serrata formed a single clade with CBS 11776, the type strain of T. yokohamensis (Fig. 8A), Kwoniella shandongensis KHU20230328-02 (GenBank: OR791286) obtained from Malus sieboldii formed a single clade with CBS 12478, the type strain of K. shandongensis (Fig. 9A). Kwoniella newhampshirensis KHU20230809-01 (Genbank: OR791285) from Ligustrum japonicum produced a single clade with CBS 13917 the holotype strain of K. newhampshirensis (Fig. 10A). Aureobasidium proteae KHU20230809-02 (Genbank: OR791281) from Ipomoea nil produced a single clade with CBS 111973, an epitype strain of A. proteae (Fig. 11A), and Fellomyces penicillatus KHU20230809-03 (Genbank: OR791283) from Zelkova serrata produced a single clade with F. penicillatus CBS 5492 (Fig. 12A). The yeasts were morphologically characterized and 12 yeasts are listed in the NSLK database.
Species description
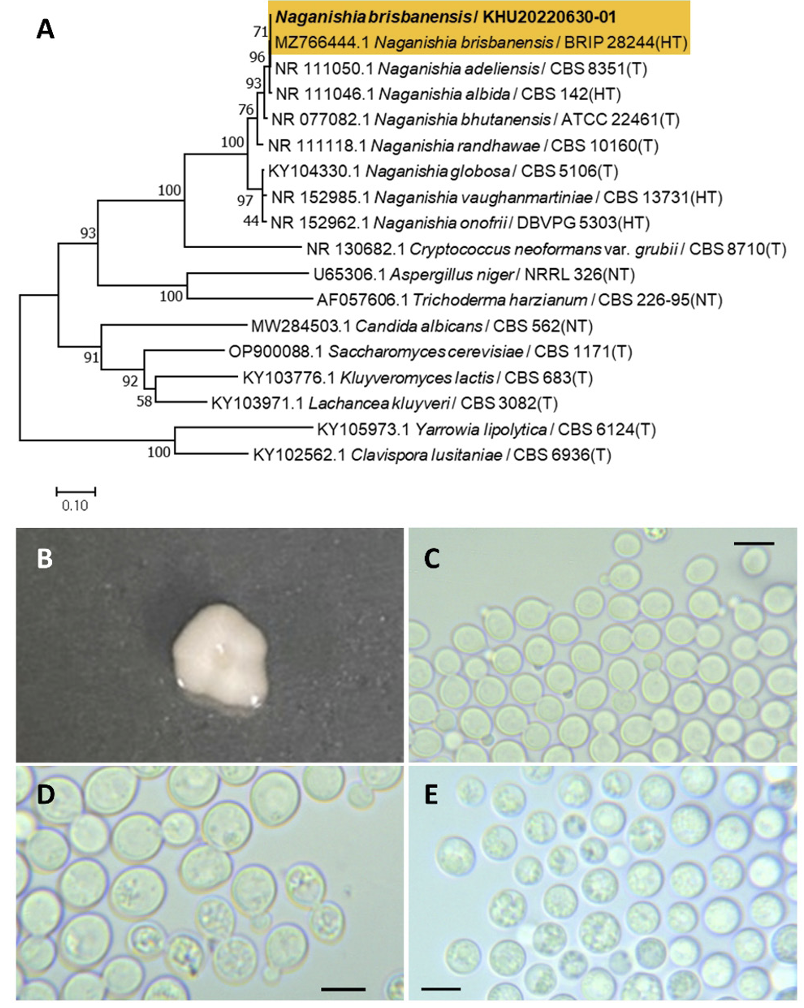
Naganishia brisbanensis Tan, Y. P., Marney, T. S., Shivas, R. G., Index Fungorum 495:4, 2021
Naganishia is a fungal genus that belongs to the Filobasidiales order. The genus Naganishia was initially described by Goto et al. (1963) [13]. Naganishia brisbanensis was distinguished from Naganishia albida [14].
Colonies were regular in shape with an entire margin and ivory to white color after 3 d on YM agar at 25℃. Cells were ovoid to elongated with a size of 7.84×7.14 μm, and occurring singly or in pairs after 3 days on YM agar at 25℃. The budding was polar and on a narrow base. A teliospore formed after two weeks of culture on Dalmau plates at 25℃ (Fig. 1B-E).
Remarks: Naganishia brisbanensis is unstudied and reported. Naganishia albida is genetically close to Naganishia brisbanensis and can produce lipids using agricultural and industrial wastes [15].
Fig. 1
Phylogenetic tree and morphological characteristics of Naganishia brisbanensis. A. Phylogenetic tree drawn from neighbor-joining analysis based on sequence of the internal transcribed spacer region, showing positions of N. brisbanensis strains isolated from Korea. Bold means representative strain. B-E. Morphology of N. brisbanensis KHU20220630-01. B. Colony on yeast aalt (YM) agar after 7 d at 25℃. C. Budding cells on YM agar after 3 d at 25℃. D. Budding cells occurring in short chains on YM agar after 7 d at 25℃. E. Teliospores are formed on Dalmau plates containing cornmeal agar after 2 weeks. Scale bars, 10 µm. (T, Type strain; HT, Holotype; NT, Neotype.)
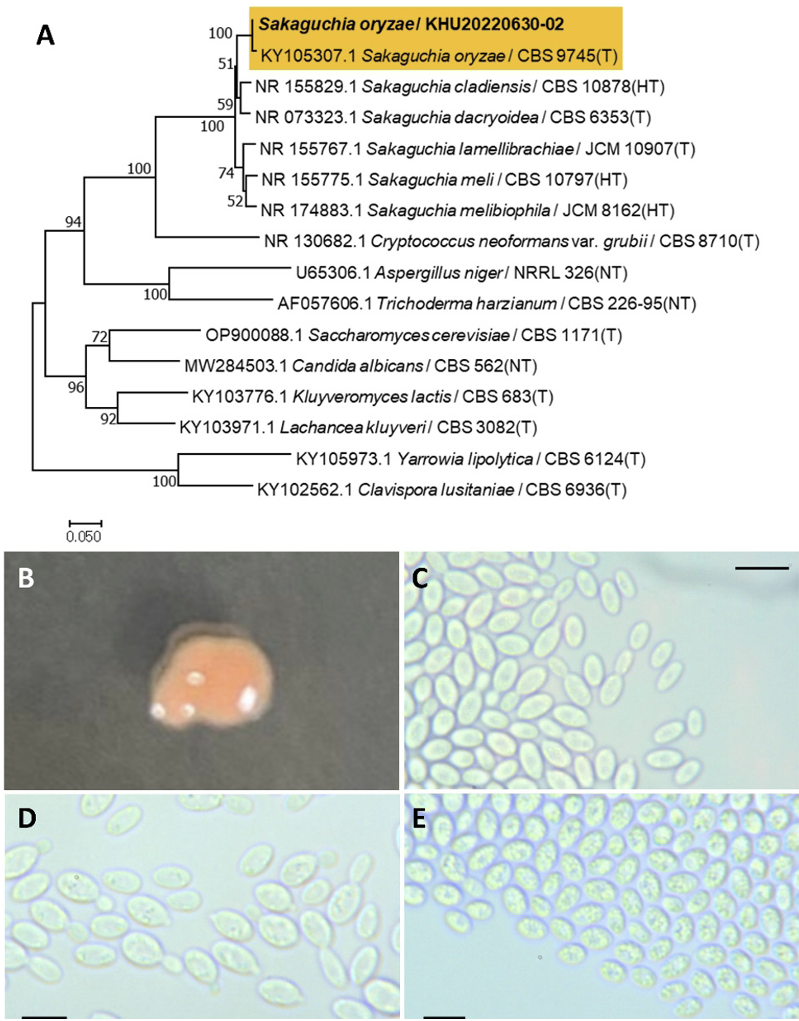
Sakaguchia oryzae (F. Y. Bai & Y. M. Cai) Wang, Q. M., Bai, F. Y., Groenewald, M., Boekhout, T., Stud. Mycol., 81(1), 149-189, 2015
Sakaguchia is a fungal genus belonging to the order Cystobasidiomycetes incertae [16]. Colonies were irregular in shape with an entire margin and orange to pink in color after 3 days on YM agar at 25℃. Cells were ovoid to elongated with a size of 9.96×4.40 μm, and occur singly or in pairs after 3 d on YM agar at 25℃. The budding was polar and on Fa narrow base. The telispore was formed after two weeks of culture on Dalmau plates at 25℃ (Fig. 2B-E). This species has teliospores lateral or terminal to the hyphae, and pseudohyphae or true hyphae present or absent [17].
Remarks: The Sakaguchia genus is unstudied, although the Rhodotorula genus related to Sakaguchia can produce carotenoids including β-carotene, and lipid can be used for biodiesel [18].
Fig. 2
Phylogenetic tree and morphological characteristics of Sakaguchia oryzae. A. Phylogenetic tree drawn from neighbor-joining analysis based on sequence of the internal transcribed spacer region, showing positions of S. oryzae strains isolated from Korea. Bold means representative strain. B-E. Morphology of S. oryzae KHU20220630-02. B. Colony on yeast malt (YM) agar after 7 d at 25℃. C. Budding cells on YM agar after 3 d at 25℃. D. Budding cells occurring in short chains on YM agar after 7 d at 25℃. E. Teliospores are formed on Dalmau plates containing cornmeal agar after 2 weeks. Scale bars, 10 µm. (T, Type strain; HT, Holotype; NT, Neotype.)
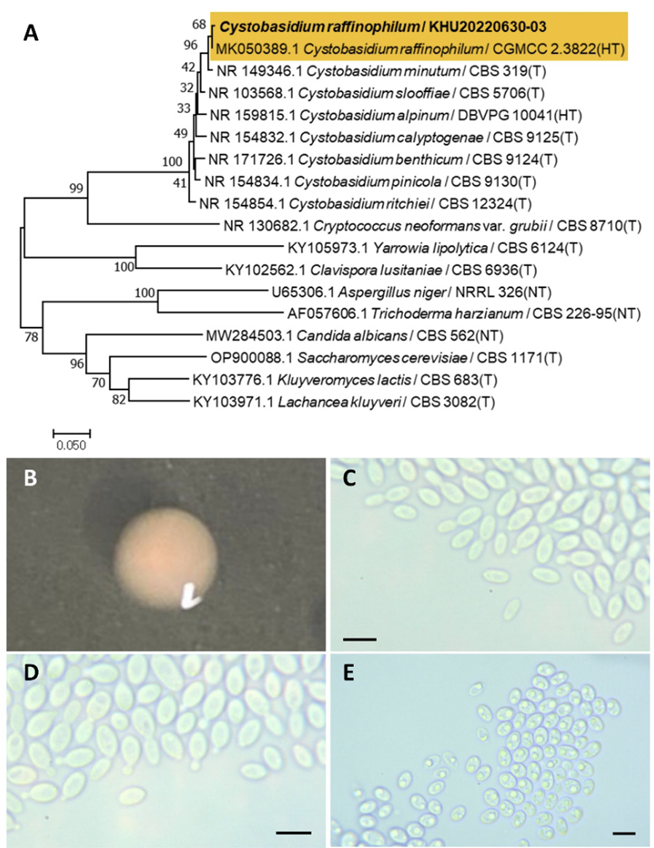
Cystobasidium raffinophilum Wang, Q. M. Bai, F. Y. Li, A. H. Stud. Mycol., 96(1), 17-140, 2020
Cystobasidium is a fungal genus belonging to the Cystobasidiales order. The genus Cystobasidium was originally established by Lagerheim in 1898 and described by Neuhoff in 1924 [19].
Colonies were round with entire margins and pink to white in color after 3 days on YM agar at 25℃. Cells were ovoid to elongated with a size of 8.85×4.20 μm, and occurred singly or in pairs after 3 d on YM agar at 25℃. The budding was polar and on a narrow base. Pseudohyphae were formed after two weeks of culture on Dalmau plates at 25℃. Ascospores were also observed (Fig. 3B-E). This species has pseudohyphae, but sexual structures were not observed [20].
Remarks: Cystobasidium raffinophilum is unstudied, although the genus Cystobasidium produces carotenoids including β-carotene, γ-carotene, torulene, and torularhodin [21]. The Cystobaisidum genus also produces lipids [22].
Fig. 3
Phylogenetic tree and morphological characteristics of Cystobasidium raffinophilum. A. Phylogenetic tree drawn from neighbor-joining analysis based on sequence of the internal transcribed spacer region, showing positions of C. raffinophilum strains isolated from Korea. Bold means representative strain. B-E. Morphology of C. raffinophilum KHU20220630-03. B. Colony on yeast malt (YM) agar after 7 d at 25℃. C. Budding cells on YM agar after 3 d at 25℃. D. Budding cells occurring in short chains on YM agar after 7 d at 25℃. E. Pseudohyphae and ascospores formed on Dalmau plates containing cornmeal agar after 2 weeks. Scale bars, 10 µm. (T, Type strain; HT, Holotype; NT, Neotype.)
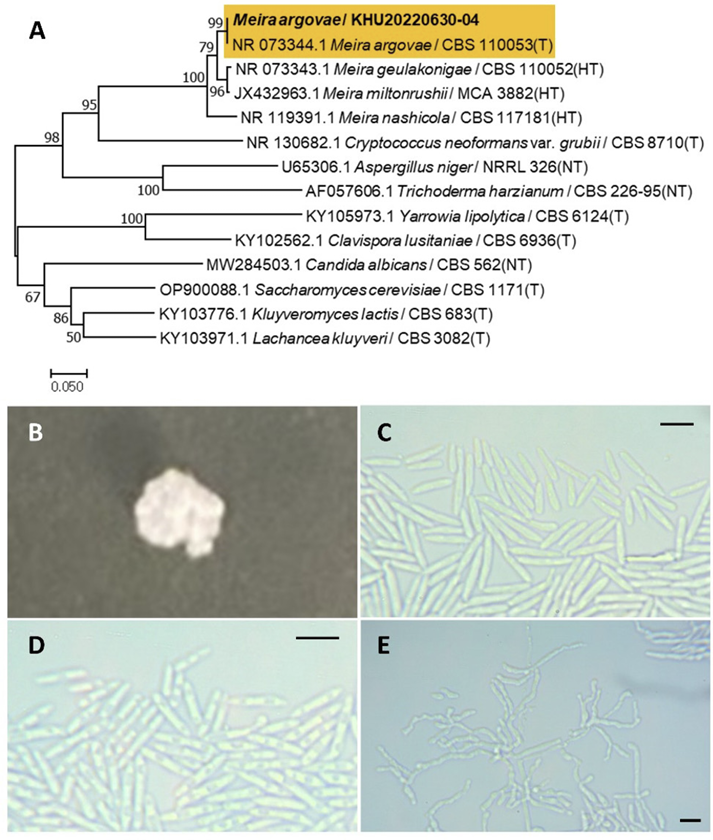
Meira argovae Boekhout, T., Scorzetti, G., Gerson, U., Sztejnberg, A., Denchev, C. M., Denchev, T. T. Mycobiota, 11, 1-10, 2021
Meira is a fungal genus that belongs to the Exobasidiales order. Meira argovae was originally described by Boekhout et al. (2003) [23] and validated by Denchev et al. (2021) owing to the invalid published name of the holotype [24].
Colonies were wrinkled with irregular margins and white in color after 3 d on YM agar at 25℃. Cells were ovoid to elongated with a size of 10.37×2.40 μm, and occurred singly or in pairs after 3 d on YM agar at 25℃. The budding was polar and on a narrow base. Hyphae were formed after two weeks of culture on Dalmau plates at 25℃ (Fig. 4B-E). This species is sympodially branched and is usually a true hyphae [23].
Remarks: Meira argovae produces a chemical compound called argovin. There are no reports on the biological activity of argovin in organisms, although it can inhibit mites or Agrobacterium tumefaciens and has the potential for use in agricultural applications [25].
Fig. 4
Phylogenetic tree and morphological characteristics of Meira argovae. A. Phylogenetic tree drawn from neighbor-joining analysis based on sequence of the internal transcribed spacer region, showing positions of M. argovae strains isolated from Korea. Bold means representative strain. B-E. Morphology of M. argovae KHU20220630-04. B. Colony on yeast malt (YM) agar after 7 d at 25℃. C. Budding cells on YM agar after 3 d at 25℃. D. Budding cells occurring in short chains on YM agar after 7 d at 25℃. E. Hyphae formed on Dalmau plates containing cornmeal agar after 2 weeks. Scale bars, 10 µm. (T, Type strain; HT, Holotype; NT, Neotype.)
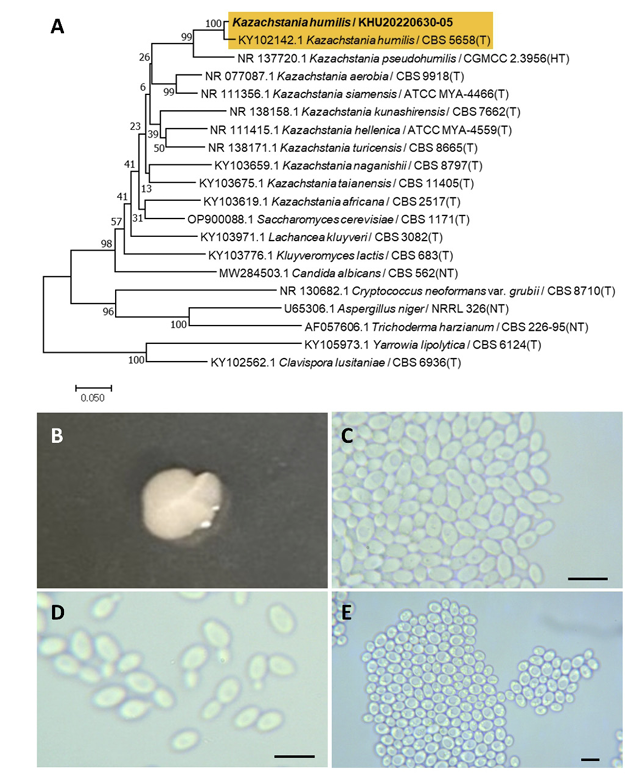
Kazachstania humilis (E. E. Nel & Van der Walt) Jacques N., Sarilar, V., Casaregola, S. Int. J. Syst. Evol. Microbiol., 66(12), 5192-5200, 2016
Kazachstania is a fungal genus belonging to the Saccharomycetales order. Kazachstania humilis has homotypic synonyms of Candida humilis. Candida humilis and Candida milleri are synonymous based on their similar nucleotide sequences [26]. However, it was later identified that Candida humilis and Candida milleri are not synonymous owing to their differences in the assimilation of sucrose and raffinose [27]. Candida humilis was reassigned as Kazachstania humilis based on sequence analysis [28].
Colonies were round with entire margins and ivory in color after 3 d on YM agar at 25℃. Cells were ovoid to elongated with a size of 6.98×4.53 μm, and occurred singly or in pairs after 3 d on YM agar at 25℃. The budding was polar and on a narrow base. Pseudohyphae were formed after two weeks of culture on Dalmau plates at 25℃. Ascospores were also observed (Fig. 5B-E). This species lacks pseudohyphae [27].
Remarks: Kazachstania humilis is a common dough-leaving agent for sourdough. This yeast is thought to be associated with other lactic acid bacteria in sourdough and cleaves dough [29].
Fig. 5
Phylogenetic tree and morphological characteristics of Kazachstania humilis. A. Phylogenetic tree drawn from neighbor-joining analysis based on sequence of the internal transcribed spacer region, showing positions of K. humilis strains isolated from Korea. Bold means representative strain. B-E. Morphology of K. humilis KHU20220630-05. B. Colony on yeast malt (YM) agar after 7 d at 25℃. C. Budding cells on YM agar after 3 d at 25℃. D. Budding cells occurring in short chains on YM agar after 7 d at 25℃. E. Pseudohyphae and ascospores formed on Dalmau plates containing cornmeal agar after 2 weeks. Scale bars, 10 µm. (T, Type strain; HT, Holotype; NT, Neotype.)
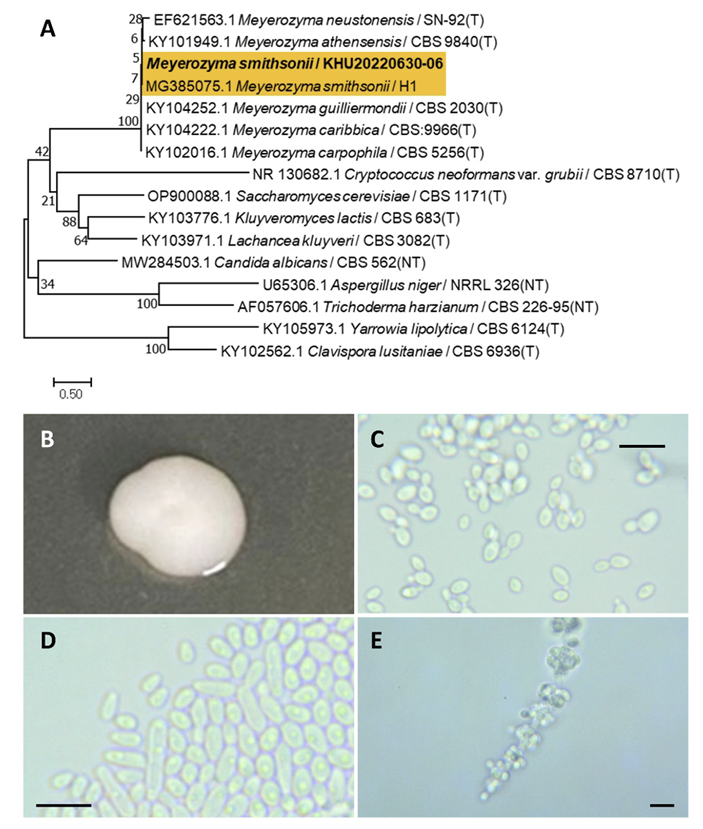
Meyerozyma smithsonii (S. O. Suh and Blackw.) A. M. Yurkov and G. Peter, Int. J. Syst. Evol. Microbiol., 67(10), 3977-3981, 2017
Meyerozyma is a fungal genus belonging to the Saccharomycetales order. Meyerozyma was originally described by Kurtzman et al., 2010 [30]. Candida smithsonii was identified and named by Suh et al. 2004 [31] and reassigned to Meyerozyma smithsonii by Yurkov et al. (2017) [32].
Colonies were round with entire margins and ivory to white in color after 3 d on YM agar at 25℃. Cells were ovoid to elongated with a size of 6.31×3.00 μm, and occurred singly or in pairs after 3 d on YM agar at 25℃. The budding was polar and on a narrow base. After two weeks of culture on Dalmau plates at 25℃ (Fig. 6B-E), pseudohyphae and blastoconidia were formed. This species possesses pseudohyphae with blastoconidia [31].
Remarks: Meyerozyma smithsonii is unstudied, although the Meyerozyma genus can degrade the mycotoxin patulin or use crude oil at contaminated sites [32,33].
Fig. 6
Phylogenetic tree and morphological characteristics of Meyerozyma smithsonii. A. Phylogenetic tree drawn from neighbor-joining analysis based on sequence of the internal transcribed spacer region, showing positions of M. smithsonii strains isolated from Korea. Bold means representative strain. B-E. Morphology of M. smithsonii KHU20220630-06. B. Colony on yeast malt (YM) agar after 7 d at 25℃. C. Budding cells on YM agar after 3 d at 25℃. D. Budding cells occurring in short chains on YM agar after 7 d at 25℃. E. Pseudohyphae and blastoconidia formed on Dalmau plates containing cornmeal agar after 2 weeks. Scale bars, 10 µm. (T, Type strain; NT, Neotype.)
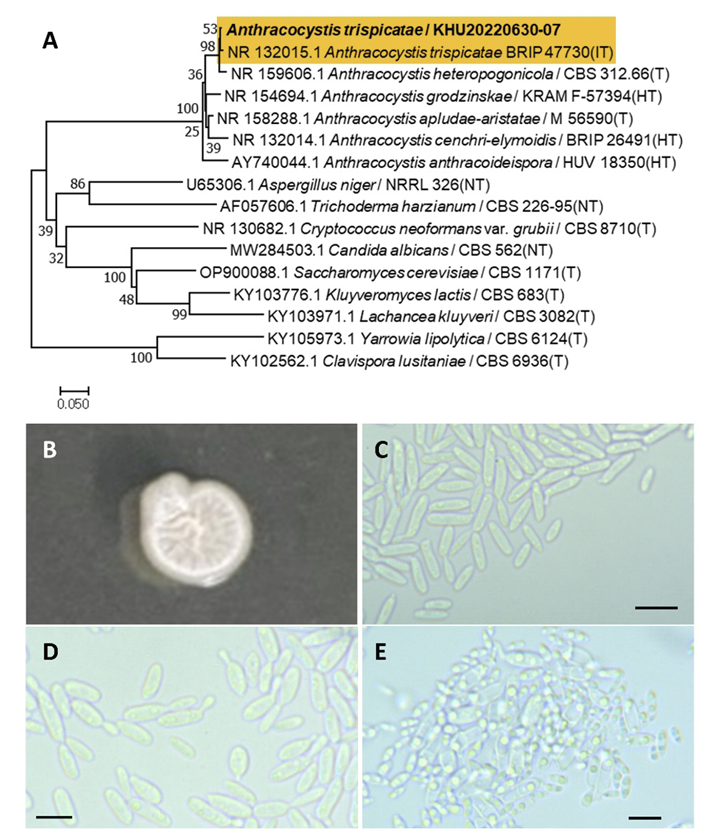
Anthracocystis trispicatae (R. G. Shivas, Vanky & Athipunyakom) McTaggart, A., Shivas, R., Persoonia, 29(1), 116-132, 2012
Anthracocystis is a fungal genus belonging to the Ustilaginales order. Anthracocystis trispicatae was reassigned by McTaggart et al., 2012 [34], and has a basionym for Sporisorium trispicatae [35] and a homotypic synonym for Sporisorium vermiculum [36].
Colonies were wrinkled and ivory to white in color after 3 d on YM agar at 25℃. Cells were ovoid to elongated with a size of 13.38×3.40 μm, and occurred singly or in pairs after 3 d on YM agar at 25℃. The budding was polar and on a narrow base. Hyphae were formed after two weeks of culture on Dalmau plates at 25℃. Furthermore, ascospores were observed (Fig. 7B-E). This species has subglobose outer spores and globoid or slightly irregular inner spores [35].
Remarks: Anthracocytis trispicatae is unstudied, although Anyhracocytis fluoculossa is genetically close to Anthracocytis trispicatae and is known for its potential against Fusarium head blight in wheat [37].
Fig. 7
Phylogenetic tree and morphological characteristics of Anthracocystis trispicatae. A. Phylogenetic tree drawn from neighbor-joining analysis based on sequence of the internal transcribed spacer region, showing positions of A. trispicatae strains isolated from Korea. Bold means representative strain. B-E. Morphology of A. trispicatae KHU20220630-07. B. Colony on yeast malt (YM) agar after 7 d at 25℃. C. Budding cells on YM agar after 3 d at 25℃. D. Budding cells occurring in short chains on YM agar after 7 d at 25℃. E. Hyphae and ascospores are formed on Dalmau plates containing cornmeal agar after 2 weeks. Scale bars, 10 µm. (T, Type strain; HT, Holotype; NT, Neotype; IT:Isotype.)
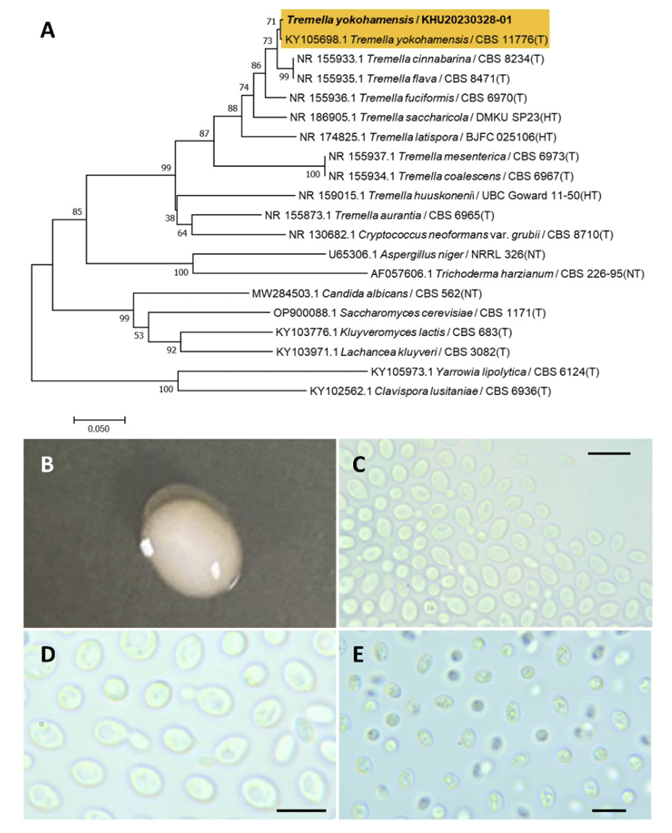
Tremella yokohamensis (Alshahni, Satoh & Makimura) Malysheva, V. F., Malysheva, E. F., Bulakh, E. M., Phytotaxa, 238(1), 40, 2015
Tremella is a fungal genus belonging to the Tremellales order. Cryptococcus yokohamenesis was originally identified by Alshahni et al. (2011) [38] and reassigned by Malysheva et al. (2015) [39] based on micromorphological features.
Colonies were round with entire margins and ivory to white in color after 3 d on YM agar at 25℃. Cells were ovoid to elongated with a size of 7.72×5.21 μm, and occurred singly or in pairs after 3 d on YM agar at 25℃. The budding was polar and on a narrow base. After two weeks of culture on Dalmau plates at 25℃, teliospores were formed (Fig. 8B-E). This species has internal hyphae and irregular septa [39].
Remarks: Tremella yokohamensis is unstudied, although the Tremella genus produces polysaccharides. Tremella polysaccharides have a unique structure and activity with applications in the food, chemical, and medicinal industries [40].
Fig. 8
Phylogenetic tree and morphological characteristics of Tremella yokohamensis. A. Phylogenetic tree drawn from neighbor-joining analysis based on sequence of the internal transcribed spacer region, showing positions of T. yokohamensis strains isolated from Korea. Bold means representative strain. B-E. Morphology of T. yokohamensis KHU20230328-01. B. Colony on yeast malt (YM) agar after 7 d at 25℃. C. Budding cells on YM agar after 3 d at 25℃. D. Budding cells occurring in short chains on YM agar after 7 d at 25℃. E. Teliospores are formed on Dalmau plates containing cornmeal agar after 2 weeks. Scale bars, 10 µm. (T, Type strain; HT, Holotype; NT, Neotype.)
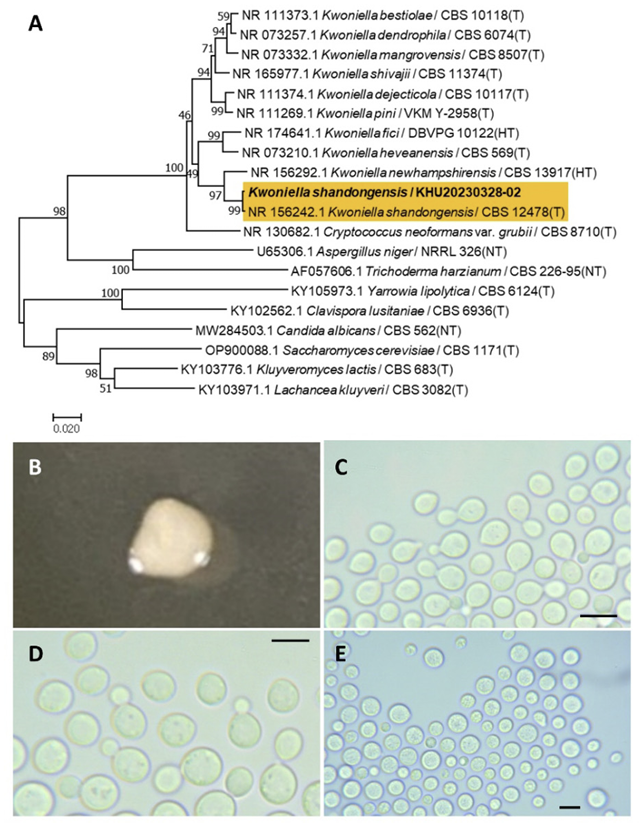
Kwoniella shandongensis Chen, R., Yuan, F. X., Jiang, M., Wei, S. C., Groenewald, M., Wang, Q. M., Stud. Mycol., 96(1), 17-140, 2020
Kwoniella is a fungal genus belonging to the Tremellales order. Kwoniella. shandongensis was first named by Chen et al. (2012) [41] by phylogenetic analysis. Li et al. (2020) validated Kwoniella shandongensis [20].
Colonies were round with entire margins and ivory in color after 3 d on YM agar at 25℃. Cells were ovoid to elongated with a size of 9.44×6.96 μm, and occurred singly or in pairs after 3 d on YM agar at 25℃. The budding was polar and on a narrow base. Telispores were formed after two weeks of culture on Dalmau plates at 25℃ (Fig. 9B-E). This species has no hyphae or sexual structures [41].
Remarks: Kwoniella shandongensis is poorly studied much, although different sequences of DNA polymerase A-related species are reported [42].
Fig. 9
Phylogenetic tree and morphological characteristics of Kwoniella shandongensis. A. Phylogenetic tree drawn from neighbor-joining analysis based on sequence of the internal transcribed spacer region, showing positions of K. shandongensis strains isolated from Korea. Bold means representative strain. B-E. Morphology of K. shandongensis KHU20230328-02. B. Colony on yeast malt (YM) agar after 7 d at 25℃. C. Budding cells on YM agar after 3 d at 25℃. D. Budding cells occurring in short chains on YM agar after 7 d at 25℃. E. Teliospores are formed on Dalmau plates containing cornmeal agar after 2 weeks. Scale bars, 10 µm. (T, Type strain; HT, Holotype; NT, Neotype.)
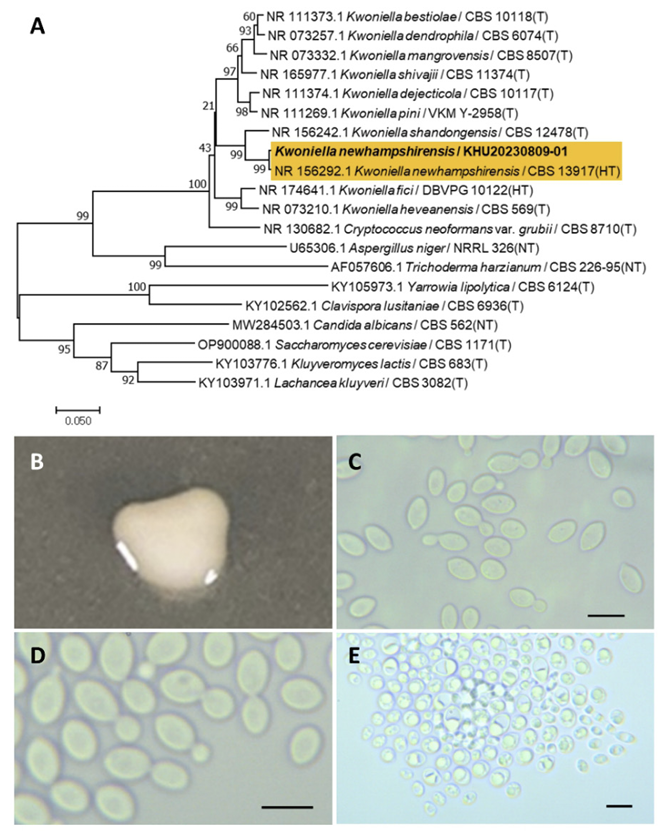
Kwoniella newhampshirensis Sylvester, K., Wang, Q. M., Hittinger, C. T., FEMS Yeast Res., 15(3), 7, 2015
Kwoniella is a fungal genus belonging to the Tremellales order. Sylvester et al. (2015) discovered Kwoniella newhampshirensis using phylogenetic analysis [43].
Colonies were round with entire margins and ivory in color after 3 d on YM agar at 25℃. Cells were ovoid to elongated with a size of 9.84×6.40 μm, and occurred singly or in pairs after 3 d on YM agar at 25℃. The budding was polar and on a narrow base. Ascospores were formed after two weeks of culture on Dalmau plates at 25℃ (Fig. 10B-E). This species was reported as ballistocondia and is not closely related to Kwoniella shandongensis [43].
Remarks: Kwoniella newhampshirensis is unstudied. However, Kwoniella produces quorum sensing inhibitors that can benefit from hybrid bioprocess optimization [44].
Fig. 10
Phylogenetic tree and morphological characteristics of Kwoniella newhampshirensis. A. Phylogenetic tree drawn from neighbor-joining analysis based on sequence of the internal transcribed spacer region, showing positions of K. newhampshirensis strains isolated from Korea. Bold means representative strain. B-E. Morphology of K. newhampshirensis KHU20230809-01. B. Colony on yeast malt (YM) agar after 7 d at 25℃. C. Budding cells on YM agar after 3 d at 25℃. D. Budding cells occurring in short chains on YM agar after 7 d at 25℃. E. Ascospores are formed on Dalmau plates containing cornmeal agar after 2 weeks. Scale bars, 10 µm. (T, Type strain; HT, Holotype; NT, Neotype.)
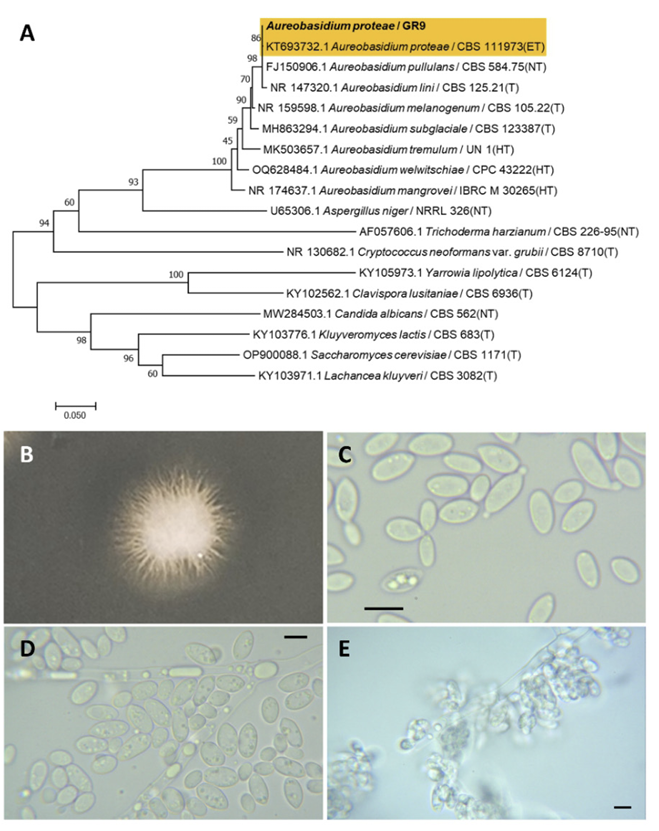
Aureobasidium proteae (Joanne E. Taylor & Crous) Taylor, J. E., Crous, P. W., Persoonia, 27(1), 20-45, 2011
Aureobasidium is a fungal genus belonging to the Dothideales order. Aureobasidium protease was originally identified as a Kabatiella protease by Taylor et al. (2000) [45]; however, the older generic name Aureobasidium is preferred over the younger and less known Kabatiella [46].
Colonies were round with radiating margins and ivory in color after 3 d on YM agar at 25℃. Cells are ovoid to elongated with a size of 15.92×5.46 μm and occurred singly or in pairs after 3 d on YM agar at 25℃. Budding was bipolar with a narrow base. Pseudohyphae and blastoconidia formed after two weeks of culture on Dalmau plates at 25℃ (Fig. 11B-E). This species is prominently constricted at the median septum with globose mucoid caps and ascospores observed [46].
Remarks: Aureobaisidum protease are pathogens responsible for chronic fungal meningitis in humans [47] and leaf blight in flowers [46].
Fig. 11
Phylogenetic tree and morphological characteristics of Aureobasidium proteae. A. Phylogenetic tree drawn from neighbor-joining analysis based on sequence of the internal transcribed spacer region, showing positions of A. proteae strains isolated from Korea. Bold means representative strain. B-E. Morphology of A. proteae KHU20230809-02. B. Colony on yeast malt (YM) agar after 7 d at 25℃. C. Budding cells on YM agar after 3 d at 25℃. D. Budding cells occurring in short chains on YM agar after 7 d at 25℃. E. Pseudohyphae and blastoconidia are formed on Dalmau plates containing cornmeal agar after 2 weeks. Scale bars, 10 µm. (T, Type strain; ET, Epitype; HT, Holotype; NT, Neotype.)
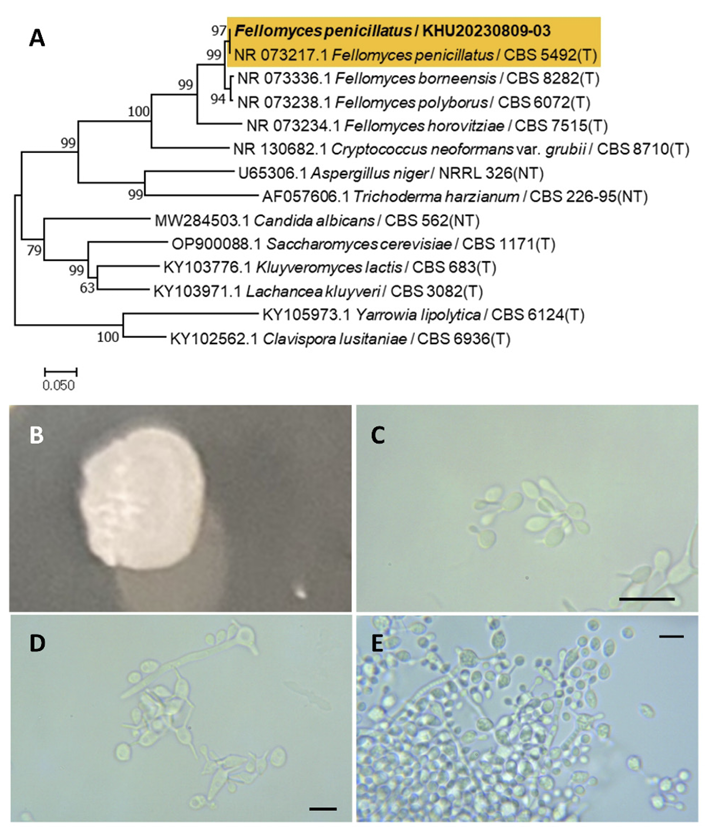
Fellomyces penicillatus (Rodr. Mir.) Yamada, Y., Banno, I., J. gen. appl. Microbiol., 30(6), 524, 1984
Fellomyces is a fungal genus belonging to the Tremellales order. Fellomyces penicillatus was first reported by Yamada et al. (1984) [48]. Before this, it was named Sterigmatomyces penicillatus by Miranda et al. (1975) [49].
Colonies were wrinkled with entire margins and ivory to white in color after 3 d on YM agar at 25℃. Cells were ovoid to elongated with a size of 5.83×4.23 μm, and occurred singly or in pairs after 3 d on YM agar at 25℃. The bud type is hypha. Hyphae were formed after two weeks of culture on Dalmau plates at 25℃ (Fig. 12B-E). A previous study did not observe hyphae and the cellular globose is ovoid [48].
Remarks: Fellomyces penicillatus is unstudied, although the Fellomyces genus is known for its long neck yeast and was used to inhibit conidiogenesis because it is rich in actin and cytoskeleton [50].
Fig. 12
Phylogenetic tree and morphological characteristics of Fellomyces penicillatus. A. Phylogenetic tree drawn from neighbor-joining analysis based on sequence of the internal transcribed spacer region, showing positions of F. penicillatus strains isolated from Korea. Bold means representative strain. B-E. Morphology of F. penicillatus KHU20230809-03. B. Colony on yeast malt (YM) agar after 7 d at 25℃. C. Budding cells on YM agar after 3 d at 25℃. D. Budding cells occurring in short chains on YM agar after 7 d at 25℃. E. Hyphae is formed on Dalmau plates containing cornmeal agar after 2 weeks. Scale bars, 10 µm (T, Type strain; NT, Neotype.)