INTRODUCTION
Soil fungi significantly contribute to ecosystems by acting as decomposers, generating diverse enzymes that facilitate the breakdown of organic matter and maintain nutrient balance. Furthermore, these key regulators in the ecosystem have a substantial influence on both plant diversity and productivity [1]. Ascomycota is the largest phylum of fungi that contains 93,000 species under 20 classes, including Eurotiomycetes, Leotiomycetes, and Sordariomycetes [1]. The phylum Ascomycota is well known for its diverse ecological characteristics and includes saprophytes, symbionts, and pathogens that are associated with soils, insects, plants, fungi, and other invertebrates [2]. The genus Chaetomella belonging to the phylum Ascomycota, class Leotiomycetes, order Chaetomellales, and family Chaetomellaceae was introduced by Fuckel in 1870 based on the production of the fruiting body pycnidium [3]. This genus was initially documented in Europe but was also observed in other regions, such as the United Kingdom and the United States. Its presence in different geographical locations indicates its adaptability and survival capabilities [4]. Morphologically, fungi belonging to this genus produce black pycnidia that generally open by a raphe; have acropleurogenous conidiogenous cells; and have nonseptate, hyaline, usually fusiform to falcate, rarely ellipsoid conidia [5]. The genus Oidiodendron belonging to the phylum Ascomycota, class Eurotiomycetes, and family Myxotrichaceae was introduced by Robak in 1932 [6]. This genus is well known as a globally distributed genus that is commonly found in various habitats, such as soil and various cellulosic substrates, and is occasionally present in lichens or the air [7]. The primary morphological attributes of this genus include upright and distinct dark conidiophores, which exhibit a high degree of branching at their upper regions to generate fertile hyphae. Moreover, these conidiophores fragment in a basipetal manner at their lower regions, resulting in the formation of arthroconidia [7]. The genus Sarocladium belonging to the phylum Ascomycota, class Sordariomycetes, order Hypocreales, and family Sarocladiaceae constitutes the soil of agricultural ecosystems [8]. This genus was first reported by Gams and Hawksworth in 1975 [9]. Several species belonging to the genus Sarocladium, including S. brachiariae and S. spinificis, are known to be saprophytic or endophytic fungi that live on Poaceae plants, such as bamboo, rice, and maize, or in the soil [10]. In addition, while some members of this genus have traditionally been regarded as significant phytopathogens, the genus also consists of opportunistic human pathogens [11]. A morphological characteristic of this genus is phialides that are one-celled, cylindrical, hyaline conidia; have a narrow cylindrical shape; hardly taper toward the apices; and lack collarettes [12]. The objectives of this study aimed to analyze the unidentified fungi isolated from soil in Korea and to determine their phylogeny and morphology. To this end, we isolated three morphologically distinct fungal strains during an investigation of unrecorded fungal species in Korea.
MATERIAL AND METHODS
Sample collection and fungal isolation
The fungal isolates used in this study were present in soil samples collected from Baekunsa, Gyeongsangbuk-do (36°27′14.14′′N, 127°56′33.40′′E); Suseo-ri, Gyeongsangbuk-do (36°11′31.4′′N, 128° 33′29.5′′E); and Gapjangsa, Gyeongsangbuk-do (36°20′54.30′′N, 128°10′38.23′′E) in Korea. Soil samples were collected from the field at a depth of 15-30 cm using a pre-autoclaved sterile spatula, air-dried, and stored at 4℃ in a plastic bag. For isolation, the soil samples (1 g) were suspended in 10 mL of sterile distilled water, vortexed gently, and serially diluted and a volume of 100 μL was spread on potato dextrose agar (PDA; Difco, Detroit, MI, USA) plates and incubated for 2-3 days at 25℃ [13]. Germinating single colonies were then transferred to new PDA plates at 25℃. A pure culture of the strain was selected for further molecular analyses based on specific characteristics. The isolates KNUF-22-003, KNUF-20-NI016, and KNUF-22005 were chosen for additional molecular analyses and cultural and morphological characterization. These isolates have been deposited at the National Institute of Biological Resources (NIBR) under the accession numbers NIBRFGC000509830, NIBRFGC000507847, and NIBRFGC000509831, respectively.
Cultural and morphological characterization
Cultural and morphological characteristics of the three fungal strains were assessed using cultural media based on the genus by following previous studies [4,7,12]. All three strains were cultured on PDA for 14 days at 25℃. KNUF-20-NI016 was also grown on oatmeal agar (OA; Difco, Detroit, MI, USA) for 14 days at 25℃, while KNUF-22-005 was also grown on cornmeal agar (CMA; Difco, Detroit, MI, USA) for 28 days at 25℃. The growth of the fungi was quantified, and details of the colonies, including their color, shape, and size, were noted. A light microscope (BX-50; Olympus, Tokyo, Japan) was used to study the morphological properties.
Genomic DNA extraction, PCR amplification, and sequencing
For the molecular identification of strains KNUF-22-003, KNUF-22-005, and KNUF-20-NI016, total genomic DNA was extracted using the HiGene™ Genomic DNA Prep Kit for fungi (Biofact, Daejeon, Korea). The internal transcribed spacer (ITS) region of the total genomic DNA extracted from the samples was amplified using ITS1F/ITS4 primer pairs [14,15], and the large subunit (LSU) region of 28S rDNA was amplified using LROR/LR5 primer pairs [16]. The actin (ACT1) gene was amplified using ACT1/ ACT4 primer pairs [17]. Amplification was confirmed by electrophoresis on 0.7% agarose gels stained with ethidium bromide. The amplification products were purified using ExoSAP-IT (Thermo Fisher Scientific, Waltham, MA, USA) and sequenced by Bioneer Co. (Daejeon, Korea).
Phylogenetic analyses
Sequences obtained through sequencing were compared for similarity using the Basic Local Alignment Search Tool (BLAST) in the National Center for Biotechnology Information (NCBI) database (Table 1). Phylogenetic trees were constructed based on combined sequences of ITS regions, LSU genes, and ACT1 genes using the neighbor-joining (NJ) method [18] in MEGA version X [19]. The evolutionary distance matrices for the NJ analysis were generated according to Kimura’s two-parameter model with bootstrap values based on 1,000 replications [20].
RESULTS AND DISCUSSION
Chaetomella oblonga Fuckel, Fungi Rhenani Exsiccati. Supplementi Fasc. 5: no. 1962 (1867) [MB#163639]
Morphology of strain KNUF-22-003
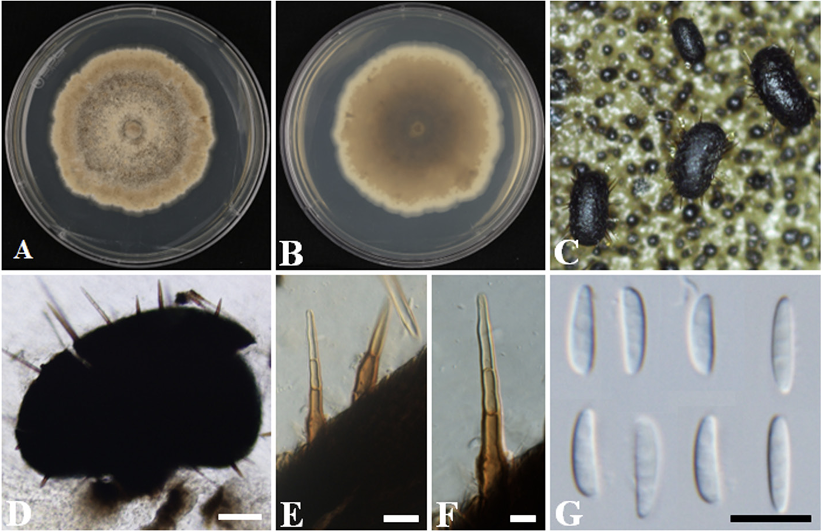
The colonies achieved a growth with a diameter of 72-81 mm in 14 days at 25℃ on PDA. The obverse appeared smooth, with a low aerial, pale vinaceous to vinaceous buff surface. The margin was white and even, having sparse wrinkles (Fig. 1A). On the reverse side, the colony appeared cream in color near the edge and darkened to dark brown in the center (Fig. 1B). The pycnidia were about 180-550×130-260 µm (n=20) and appeared to be elongated, oblong, rarely globose, and dark (Fig. 1C and D). The setae were 106.9-320.0×7.0-10.5 µm (n=30) and dark brown, and appear paler at the apex, with shapes varying from clavate to pointed, found all over the fruiting body and sharp (Fig. 1E and F). The pycnidia developed in the culture were discoid and stalked; the stalks were pale brown and surrounded by setae. The conidia were non-septate, hyaline, and fusiform to falcate with ends that were slightly pointed, straight to curved, and smooth; with a diameter of 7.7-10.9×1.5-2.7 μm (n=50) (Fig. 1G). The cultural and morphological characteristics of strain KNUF-22-003 indicate that it is most closely related to C. oblonga 110.78T [4].
Fig. 1
Cultural and morphological characteristics of strain KNUF-22-003. Cultures were grown at 25℃ on potato dextrose agar (PDA). A, B: Front and reverse views of the colony after 14 days; C: Pycnidia in culture; D: Conidia released from pycnidia; E, F: Setae on pycnidia; G: Conidia. Scale bars: D=100 µm; E-G=10 µm.
Molecular phylogeny of strain KNUF-22-003
Based on sequencing analyses, the length of the sequences obtained from the strain KNUF-22-003 for the ITS regions and LSU gene, was 483 and 812 bp, respectively. The ITS region showed 100% similarity Based on sequencing analyses, the length of the sequences obtained from the strain KNUF-22-003 for the ITS regions and LSU gene, was 483 and 812 bp, respectively. The ITS region showed 100% similarity to the C. oblonga strains BPI 843552 and BPI 843553. The LSU gene sequences of the strain showed 99.9% similarity to the strains C. oblonga BPI 843552 and BPI 843553 and 98.6% similarity to the strain C. oblonga CBS 110.78T. The NJ phylogenetic tree (a combination of ITS region and LSU gene sequences; Fig. 2) indicated that strain KNUF-22-003 was clustered with various strains of C. oblonga (CBS 110.78T, BPI 843552, and BPI843553). Thus, based on morphological and phylogenetic analyses, strain KNUF-22003 was identified as C. oblonga, a newly described fungus in Korea.
Currently, about 40 species in the genus Chaetomella have been reported worldwide. They are saprophytes growing on litter, dead logs, and soil distributed in both temperate and tropical regions [21]. Moreover, C. raphigera has been reported as the causal agent of leaf spot disease in various hosts, including cigar flower (Cuphea spp.) [22]. Since C. raphigera is the only species in the genus Chaetomella reported in Korea [23], that this report of C. oblonga will contribute to the study of Chaetomella taxa in Korea.
Fig. 2
Neighbor-joining phylogenetic tree based on a combined dataset of partial sequences of internal transcribed spacer (ITS) regions and partial large subunit (LSU) gene sequences showing the phylogenetic position of strain KNUF-22-003 among Chaetomella species and its closest relationship with Chaetomella oblonga. Bootstrap values greater than 70% (percentage of 1,000 replications) are shown at branching points. The strain isolated in this study is denoted in bold and red. The tree was rooted using Glomerella cingulata AR3788 as an out-group. Bar, 0.02 substitutions per nucleotide position. T indicates type strain.
Oidiodendron chlamydosporicum Morrall, Canadian Journal of Botany 46: 205 (1968) [MB#335316]
Morphology of strain KNUF-22-005
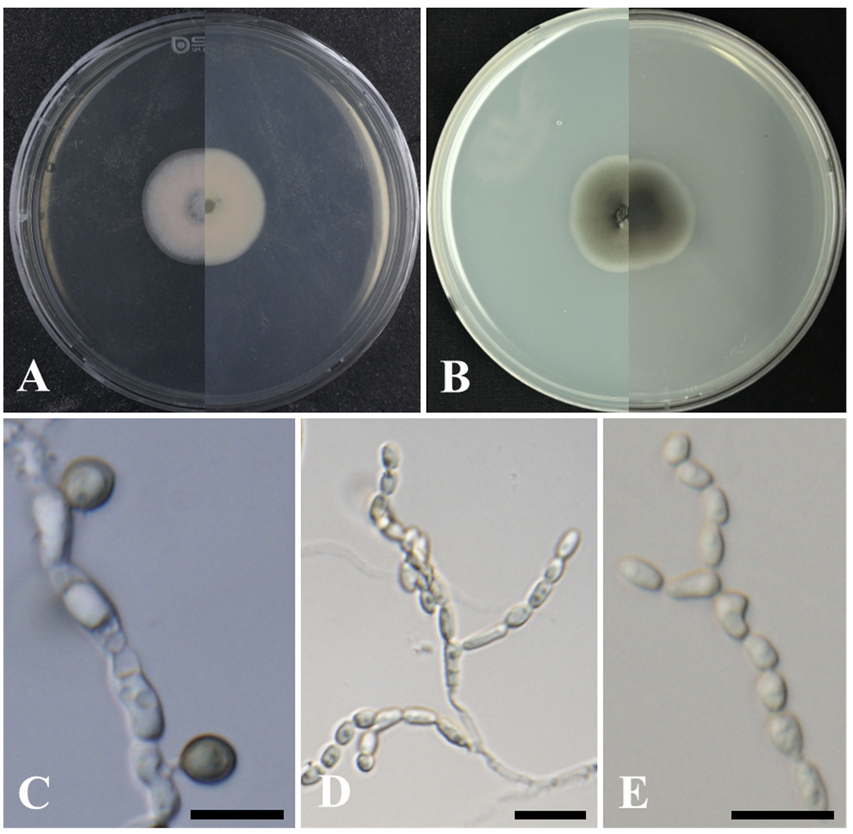
The colonies achieved growth with a diameter of 29- 31 mm in 28 days at 25℃ on PDA. The colonies were dark green to black at first but aged to a light olive gray color at the margin (Fig. 3A). The strain also achieved growth with a diameter of 17 -30 mm in 28 days at 25℃ on CMA. The colonies on CMA appeared to be cream or light gray to green–gray, or brown; with darker margins and appressed on the obverse side. However, on the reverse side, the colony appeared cream near the edge and darkened to dark brown in the center (Fig. 3B). The chlamydospores were terminal, intercalary, subglobose, and melanized with a diameter of 3.7-7.6×2.6-5.6 μm (n=30) (Fig. 3C). Conidiophores were melanized and bearing chains of hyaline conidia, with the melanized chlamydospores alternating between short, branched, and lightly pigmented to melanized structures and erect structures (Fig. 3D). The conidia were thin-walled, globose, subglobose, or elongated with a diameter of 2.5-5.5×1.4-3.0 μm (n=30) (Fig. 3E). The cultural and morphological characteristics of strain KNUF-22-005 indicate that it is most closely related to O. chlamydosporicum CBS 403.69T [7].
Fig. 3
Cultural and morphological characteristics of strain KNUF-22-005. Cultures were grown at 25℃ for 28 days. A, B: Front and reverse views on potato dextrose agar (PDA) and cornmeal agar (CMA), respectively; C: Chlamydospore; D, E: Elongate, hyaline conidia produced in short chains at the apices of short and dark conidiophores. Scale bars: C-E=10 µm.
Molecular phylogeny of strain KNUF-22-005
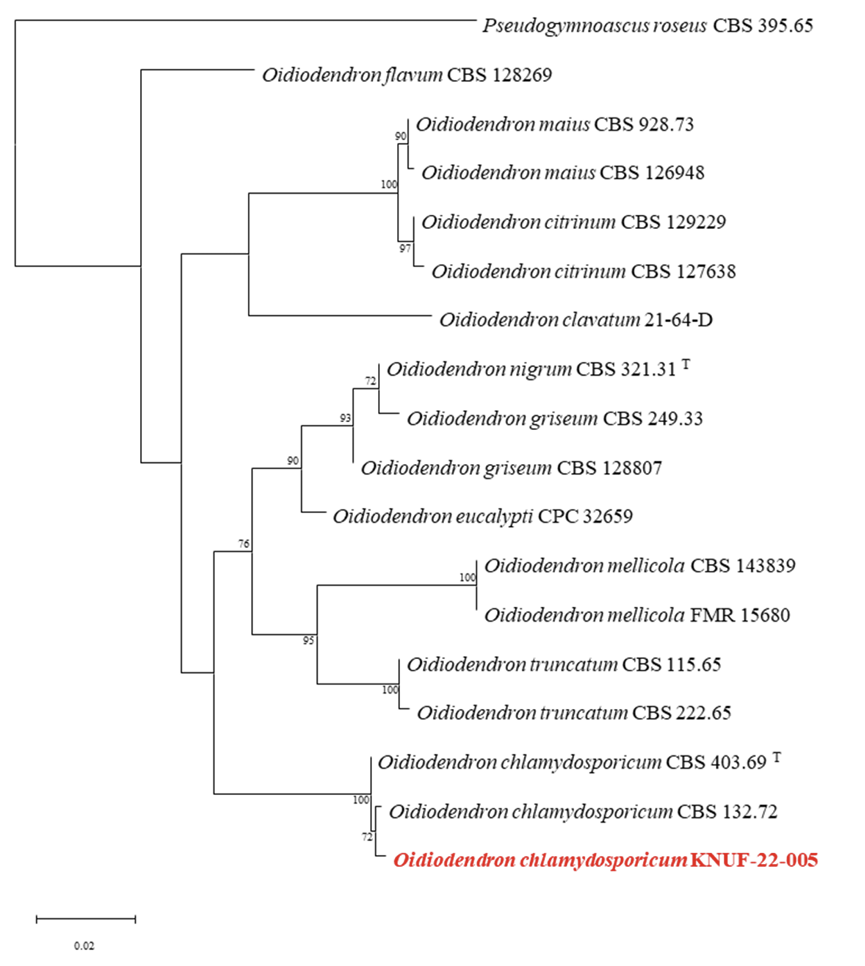
Based on sequencing analyses, the length of the sequences obtained from the strain KNUF-22-005 for the ITS region and LSU gene, was 459 and 810 bp respectively. The ITS region showed 99.6% and 99.6% similarity to the strains O. chlamydosporicum CBS 403.69T and 132.72, respectively. The LSU gene sequences of the strain showed 99.6% similarity to the strains O. chlamydosporicum CBS 403.69T, and CBS 132.72. The NJ phylogenetic tree (a combination of ITS regions and LSU gene sequences; Fig. 4) indicated that strain KNUF-22-005 was clustered with previously identified strains of O. chlamydosporicum (Fig. 4). Based on morphological and phylogenetic analysis, strain KNUF-22-005 was identified as O. chlamydosporicum, a newly described fungus in Korea.
Of the 32 species belonging to the genus Oidiodendron, only 5 species (O. citrinum, O. echinulatum, O. f lavum, O. maius, and O. clavatum) have been reported in Korea [24]. O. maius is a mycorrhizal fungus that facilitates nutrient exchange in plants and makes plants tolerant to various metals, such as Zn and Cd, thereby affecting plant growth and nutrient movement [25]. The genus Oidiodendron, which can have direct and indirect effects on plants, can also have a significant impact on humans. Chetracin B, a secondary metabolite produced by O. truncatum, is cytotoxic against human cancer cells [26]. This suggests that Oidiodendron exhibits both medicinal and pathogenic effects and that precautions should be taken against its pathogenicity. The genus Oidiodendron is a mycorrhizal fungus that can directly act on plants. It is being studied for its functional diversity and availability for medical purposes; however, there is still a lack of research on O. chlamydosporicum and the genus Oidiodendron as a whole.
Fig. 4
Neighbor-joining phylogenetic tree based on a combined dataset of partial sequences of internal transcribed spacer (ITS) regions and partial large subunit (LSU) gene sequences showing the phylogenetic position of strain KNUF-22-005 among Oidiodendron species. Bootstrap values greater than 70% (percentage of 1,000 replications) are shown at branching points. The strain isolated in this study is denoted in bold and red. The tree was rooted using Pseudogymnoascus roseus CBS 395.65 as an out-group. Bar, 0.02 substitutions per nucleotide position. T indicates type strain.
Sarocladium subulatum Giraldo, Gené & Guarro, Persoonia 34: 20 (2015) [MB#807948]
Morphology of strain KNUF-20-NI016
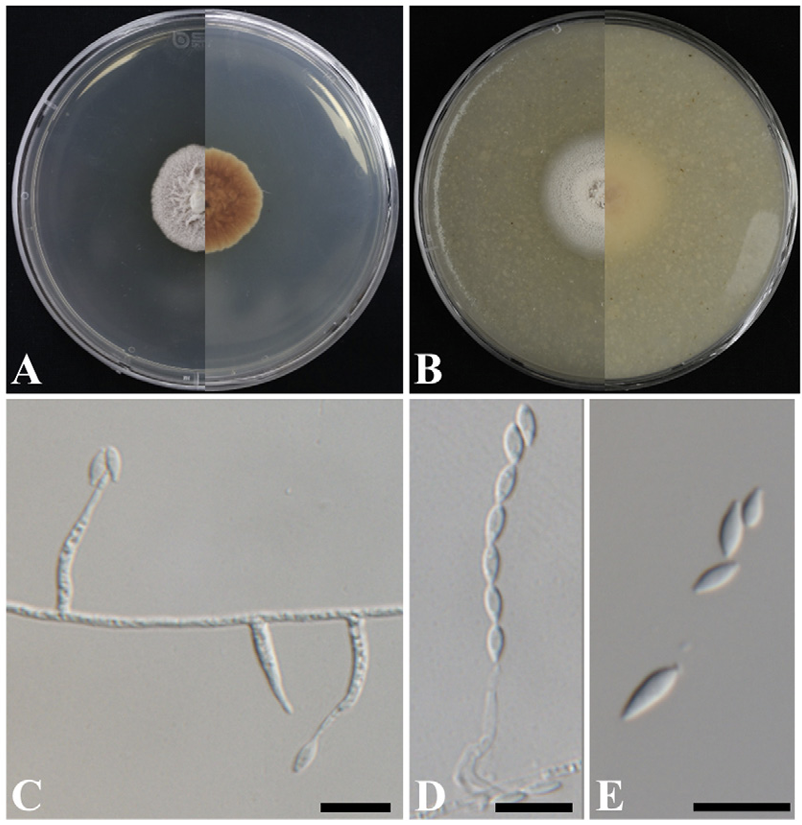
The colonies achieved growth with a diameter of 23-26 mm in 14 days at 25℃ on PDA. The obverse side appeared to be yellowish white and membranous. It became velvety and formed a crateriform shape over time (Fig. 5A). The colonies grew to 28-32 mm in 14 days at 25℃ on OA (Fig. 5B). The colonies on CMA appeared flat with a diffuse margin and were powdery with a yellowish white color. The hyphae of strain KNUF-20-NI016 were thin and transparent and had a smooth surface with septae observed. The conidiophores were erect, mostly unbranched, and simple, and were observed to be smooth and transparent (Fig. 5D). The phialides arising directly from vegetative hyphae were straight or slightly flexuous and exhibited an awl-like shape with a slender, elongated, pointed tip; 14-23 µm long and 1.4-2.6 µm wide at the base (Fig. 5C). Similar to the conidia, the hyphae were transparent and had a thin, smooth surface (Fig. 5D and E). The cultural and morphological characteristics of strain KNUF-20-NI016 indicate that it is most closely related to S. subulatum CBS 217.35T [8].
Fig. 5
Cultural and morphological characteristics of strain KNUF-20-NI016. Cultures were grown at 25℃ for 14 days. A, B: Front and reverse views on potato dextrose agar (PDA) and oatmeal agar (OA), respectively; C: Phialides arising directly from vegetative hyphae; D: Conidia arranged in chains; E: Conidia. Scale bars=10 µm.
Molecular phylogeny of strain KNUF-20-NI016
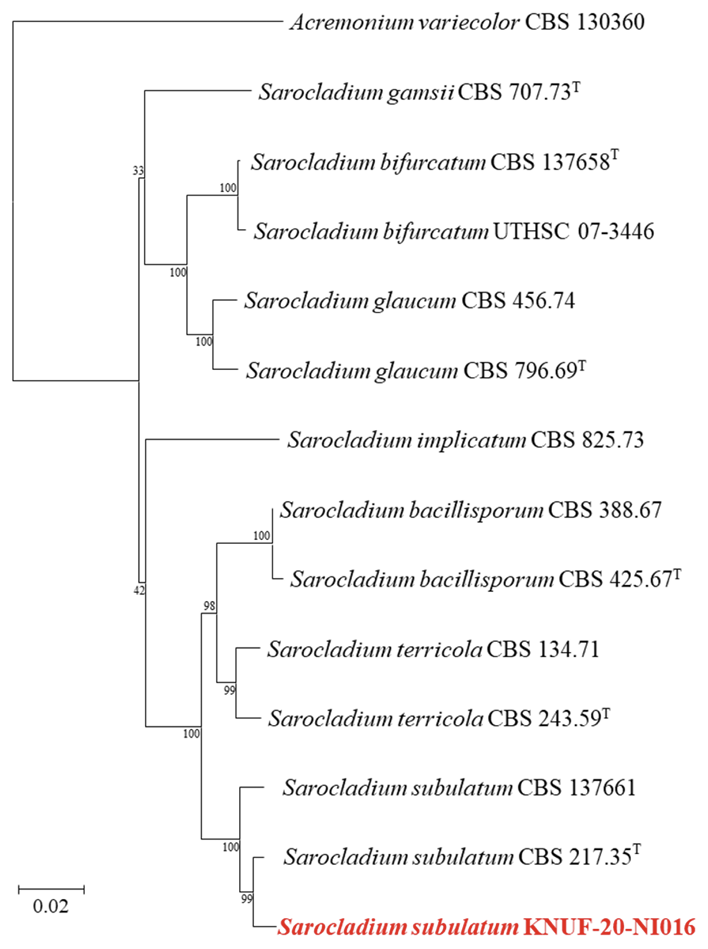
Based on sequencing analyses, the length of the sequences obtained from the strain KNUF-20-NI016 for ITS regions, LSU, and ACT1 genes, was 600, 900, and 780 bp, respectively. The ITS region showed 100% similarity to the strains S. subulatum CBS 217.35T and CBS 137661. The LSU gene sequences of the strain showed 99.9% similarity to various Sarocladium strains, such as S. bacillisporum CBS 425.67T, S. terricola CBS 243.59T, and S. subulatum CBS 217.35T. Moreover, the ACT1 gene sequences showed the highest similarity of 98.1% to S. subulatum 217.35T, followed by S. subulatum CBS 137661 (96.8% similarity) and S. bacillisporum CBS 485.67T (93.1% similarity). The NJ phylogenetic tree (a combination of ITS regions, LSU region, and ACT1 gene sequences; Fig. 6) indicated that strain KNUF-20-NI016 was clustered with previously identified strains of S. subulatum (Fig. 6). Based on morphological and phylogenetic analysis, strain KNUF-20-NI016 was identified as S. subulatum, a newly described fungus in Korea.
Several species belonging to the genus Sarocladium are traditionally known as plant pathogens [11]. For example, S. oryzae has been reported as a pathogen causing leaf blight in rice [9]. Although the pathogenicity of S. subulatum was not assessed in this study, further research is necessary considering that several fungi belonging to the genus Sarocladium act as plant pathogens and mycorrhizal fungi have also been reported as plant pathogens.
According to the effectuation of the Nagoya Protocol in 2014, knowledge of biodiversity is becoming increasingly important [27]. Accordingly, to obtain domestic biological resources and increase our knowledge of fungal diversity, 3 newly discovered species, C. oblonga, O. chlamydosporicum, and S. subulatum, were isolated from soil samples collected from Korea. The three genera are well-known causal agents of various plant pathogens on a wide range of host plants. Therefore, further research is needed for a better understanding of their diversity, geographical distribution, pathogenicity, and ecological and biological roles, particularly under Korean environmental conditions. Through this research, we believe that we can discover biological resources that can become the mainstay of the Korean bio-industry and contribute to the promotion of biodiversity and the growth of the Korean bio-industry.
Fig. 6
Neighbor-joining phylogenetic tree based on a combined dataset of partial sequences of internal transcribed spacer (ITS) regions, partial large subunit (LSU) sequences, and actin (ACT1) gene sequences showing the phylogenetic position of strain KNUF-20-NI016 among Sarocladium species and its closest relationship with Sarocladium subulatum. Bootstrap values greater than 99% (percentage of 1,000 replications) are shown at branching points. The strain isolated in this study is denoted in bold and red. The tree was rooted using Acremonium variecolor CBS 130360 as an out-group. Bar, 0.02 substitutions per nucleotide position. Tindicates type strain.