INTRODUCTION
The family Phaeosphaeriaceae was first established by Barr (1979) with the designation of Phaeosphaeria I. Miyake as the generic type of the family [1]. According to the Catalogue of Life (http://www.catalogueoflife. org; COL version 2024.02.24), this family currently comprises more than 170 genera. Notably, these fungi are both ecologically and economically significant due to their ability to adaptively shift between endophytic, pathogenic, or saprobic life strategies depending on environmental conditions [2-7]. When functioning as saprobes, the members of the family Phaeosphaeriaceae facilitate the decomposition of dead plant material. However, this family also harbors several important plant pathogens, such as Alternaria, Bipolaris, Didymella, Leptosphaeria, Neosetophoma, Parastagonospora, Phaeosphaeria, Pyrenophora, and Stemphylium species, which are responsible for severe diseases in economically significant crops [8-12]. The genus Phaeosphaeria, belonging to this family, comprises over 200 species listed in the Mycobank (https://www.mycobank.org) and Index Fungorum (http://www.indexfungorum.org) databases. Numerous species of Phaeosphaeria have been recently described, including P. blodgettiae Y.P. Tan, Bishop-Hurley, Marney & R.G. Shivas, P. chengduensis Wanas. & Maharachch., P. scalesiae Crous, P. sichuanensis Wanas. & Maharachch., and P. stonesiae Y.P. Tan, Sbaraini & E. Lacey, contributing to our knowledge of the diversity of this fungal genus [13-16]. Some species within the genus Phaeosphaeria play a crucial role as saprophytes, actively participating in processes such as decomposition and nutrient cycling. They are often associated with decaying plant material, deadwood, or organic debris from monocotyledons. Specifically, the aforementioned P. chengduensis is saprobic and was isolated from dead twigs of an unknown deciduous host. However, certain species of Phaeosphaeria are recognized as plant pathogens, causing several diseases in various hosts, including crops and forest trees [16].
Due to the remarkable diversity of microbial communities associated with insects, interactions with microbiome members can result in a wide range of effects on the fitness and behavior of insects [17]. Fungi and insects interact reciprocally through a wide array of symbiotic relationships, ranging from instances of parasitism, whereby the fungi gain an advantage by harming the insects, to those where fungi form mutualistic associations with their host [18]. The citrus flower chafer (Gametis jucunda Faldermann) is distributed across various regions such as India, Nepal, Tonkin, Taiwan, China, the former Soviet Union, Korea, Japan, and North America [19]. These insects are commonly found on flowers and tree saps across a variety of plant species [20]. Known for their harmful effects, these insects negatively impact citrus fruits, flowers, calyxes, and various other plants [21]. Adults have been observed feeding on Chinese privet (Ligustrum sinense Lour.), Wampee [Clausena lansium (Lour.) Skeels], and Orange climber [Toddalia asiatica (L.) Lam.] flowers in Macau. Additionally, in Hong Kong, they have been found on Gray nicker (Guilandina bonduc L.), True rhus (Rhus chinensis Mill), and Chinese guger tree (Schima superba Gard et Champ var. superba) [19]. Therefore, similar to other insects, Gametis jucunda could act as a potential vector for the transmission of microfungi. Nonetheless, conclusive evidence regarding the relationship between Gametis jucunda and Phaeosphaeriaceae fungi is currently lacking.
This study focused on investigating less-explored sources with the expectation of discovering previously unreported and novel fungal species. Here, we describe the identification of an insect-associated fungal strain belonging to the genus Phaeosphaeria, isolated from the citrus flower chafer (Gametis jucunda), using cultural, morphological, and molecular phylogenetic approaches.
MATERIALS AND METHODS
Collection and isolation of the fungal strain
The fungal strain utilized in this study was isolated from specimens of Gametis jucunda collected from Chopyeong-myeon, Jincheon-gun, Chungcheongbuk-do (36°49'51.7"N 127°34'08.1"E), South Korea. Following a previously described method, the fungi were isolated using potato dextrose agar (PDA, Difco, Detroit, MI), and incubated at 25℃ for 2-3 days [22]. Single colonies were individually transferred onto PDA plates and then incubated at 25℃ for 4-5 days. After being transferred onto new PDA plates and forming colonies, the grey-whitish mycelium was subsequently selected for comprehensive analysis and designated as KNUF-4H-A. The fungal isolate KNUF-4H-A was stored at -80℃ in 20% glycerol stock. The isolate, designated as KNUF-4H-A (NIBRFGC000512438), was deposited in the National Institute of Biological Resources (NIBR) as a stock culture.
Morphological characterization
To examine its cultural and morphological characteristics, the isolated fungal strain KNUF-4H-A was cultured on PDA. The characteristics of the colonies, including their color, texture, growth rates, shape, and size, were thoroughly examined after 4 weeks [16]. Additionally, morphological characteristics were observed using a BX-50 microscope (Olympus, Tokyo, Japan).
Molecular analysis
Total genomic DNA was extracted using a commercial extraction kit (HiGene Genomic DNA Prep Kit, Biofact, Daejeon, Korea) according to the manufacturer’s instructions. Polymerase chain reaction (PCR) was then performed to amplify the internal transcribed spacer (ITS) regions, using the genomic DNA as the template. The obtained amplicons were then purified using the ExoSAP-IT PCR product cleanup reagent (Thermo Fisher Scientific, Waltham, MA, USA). Recent research has successfully classified 20 species of Phaeosphaeria using the small subunit rDNA (SSU), large subunit rDNA (LSU), translation elongation factor 1-α (TEF1-α), and RNA polymerase II second largest subunit (RPB2), identifying new species such as P. chengduensis and P. sichuanensis [16]. Five loci were amplified, including the ITS, SSU, LSU, TEF1α, and RPB2 genes using the ITS1F/ITS4 [23,24], NS1/NS4 [24], LROR/LR5 [25], EF1/EF2 [26], and 5F2/7CR [27] primer pairs, respectively. The gene sequences were assembled using SeqMan software (DNASTAR, Madison, WI, USA). Next, gaps and terminal ends in the alignment were edited using BioEdit version 5.0.6 to ensure the accuracy and completeness of the gene sequences. A phylogenetic tree was constructed based on the concatenated sequences of the SSU, LSU, ITS regions, TEF1-α, and RPB2 genes via the maximum likelihood method with 1,000 bootstrap replicates using the MEGA 7 software [28]. Genetic divergence between the species was evaluated using Kimura's two-parameter model [29]. To assess evolutionary connections and genetic relatedness among Phaerosphaeria species, sequence alignments were conducted using the National Center for Biotechnology Information (NCBI) Basic Local Alignment Search Tool (BLAST). Upon conducting these BLAST searches, the closest phylogenetic sequences were identified and pairwise sequence similarity values were calculated for each gene, thereby aiding in the determination of genetic connections between Phaerosphaeria species. The gene sequences of closely related phylogenetic relatives were obtained from the NCBI GenBank database (Table 1).
RESULTS AND DISCUSSION
Morphological characteristics of the KNUF-4H-A fungal strain
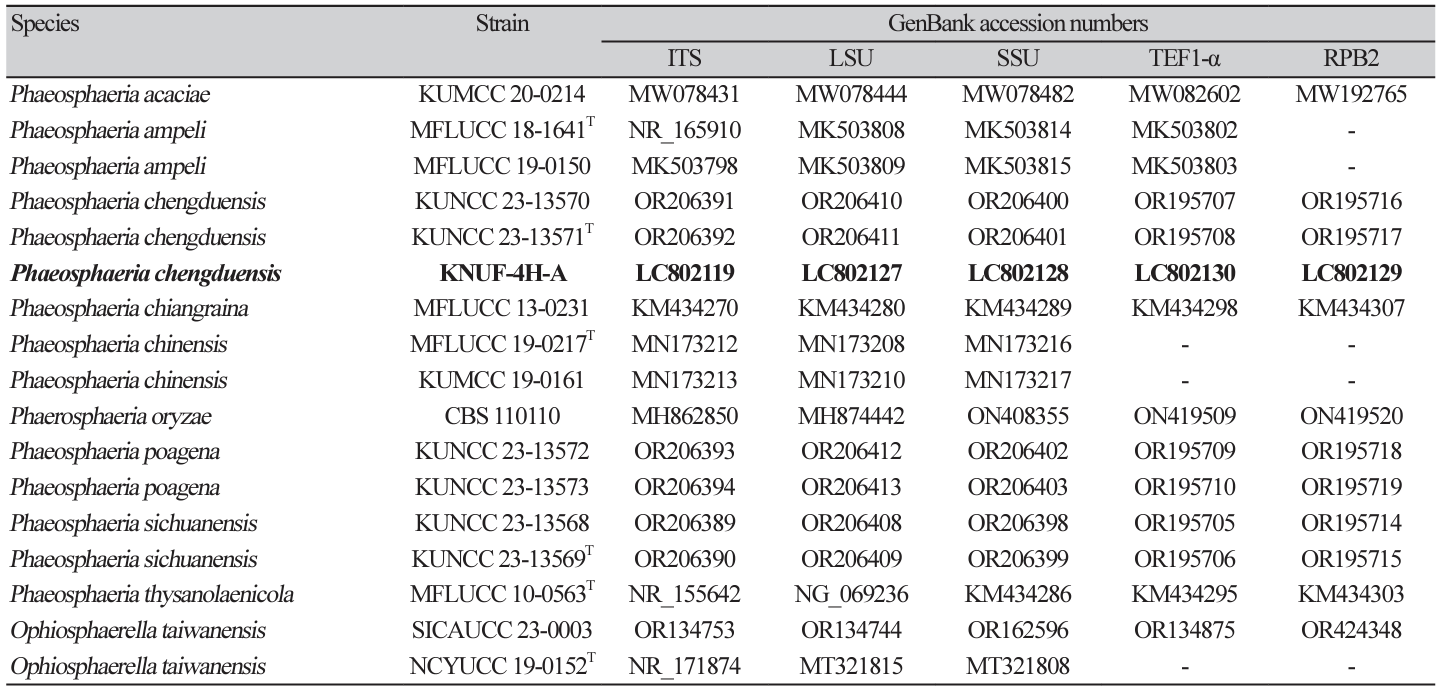
Strain KNUF-4H-A formed circular, convex, flattened, velvety, medium-dense colonies with a greenishgrey edge and a grey-whitish center. The reverse side of the colonies appeared yellowish-brown at the margin and dark brown at the center. Typically, the colonies reached a diameter of 64.4-65.1 mm after 4 weeks of growth at 25℃ on PDA medium (Fig. 1A and B). Pycnidial conidiomata were black, globose, and erumpent on the surface of the PDA media after 2 to 4 weeks (Fig. 1C). The conidiophores were borne from the inner hyphal tissue of pycnidia and produced conidia. Conidiophores were reduced to conidiogenous cells, which were hyaline, smooth, and straight to slightly curved (Fig. 1D and E). Strain KNUF-4H-A reproduced asexually through the production of conidia in conidiophores. The chlamydospores appeared hyaline, primarily arranged in chains, smooth, thick-walled, and had a globose to subglobose structure, with a diameter ranging from 4.58 to 13.92 μm (M=6.47×9.21 μm, n=25) (Fig. 1F and G). The asexual morphological characteristics of KNUF-4H-A included the formation of yellowish to olive, cylindrical conidia that were either non-septate or one-septate (Fig. 1H). The size of conidia was 3.2-8.1×2.1-3.2 μm (M=6.2×2.5 μm, n=50). The cultural characteristics of the isolate on PDA were identical to those of P. chengduensis KUNCC 23-13571T, with colonies appearing flattened, greenish-grey at the edges, greywhite at the center, and dark brown on the reverse side (Table 2). The asexual morphological characteristics of this species have not been reported. P. poagena Crous & Quaedvl. was identified as the closest neighbor of P. chengduensis according to our phylogenetic analysis and data. Therefore, P. poagena was selected as the reference species to conduct a comparative analysis of asexual morphology characteristics [16]. In contrast to KNUF-4H-A, the conidia of P. poagena CBS 136771T are solitary, brown, ellipsoidal to subcylindrical with 1-3 septa, slightly constricted at the septa, exhibiting a subobtuse apex, a truncate base, and with a notably longer length on average (Table 2). The differences in color, shape, and size of the conidia distinguish KNUF-4H-A from P. poagena, thus providing indirect evidence of its affiliation with P. chengduensis.
Fig. 1
Cultural and morphological characteristics of Phaerosphaeria chengduensis KNUF-4H-A. A, B: Front and reverse view of colony grown on potato dextrose agar (PDA) at 25℃ for 4 weeks. C: Pycnidia on PDA. D, E: Conidiophores. F, G: Chlamydospores. H: Non-septate and single-septum conidia. Scale bars: C=200 μm, D-H=10 μm.
Molecular analysis of the fungal strain KNUF-4H-A
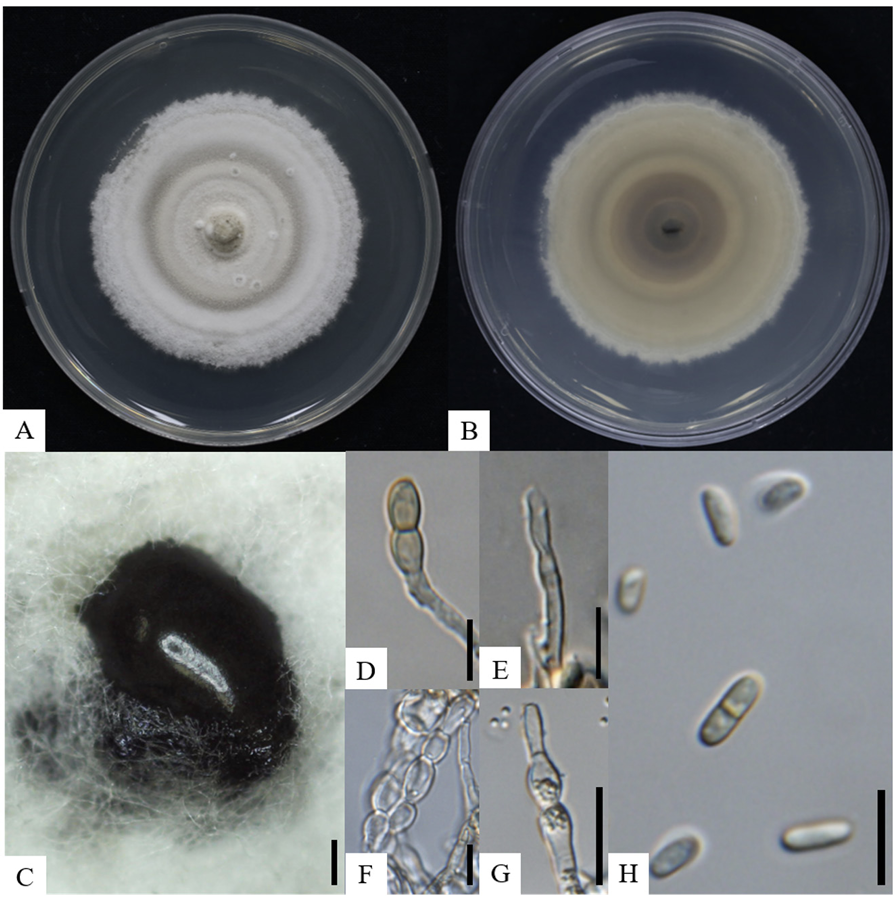
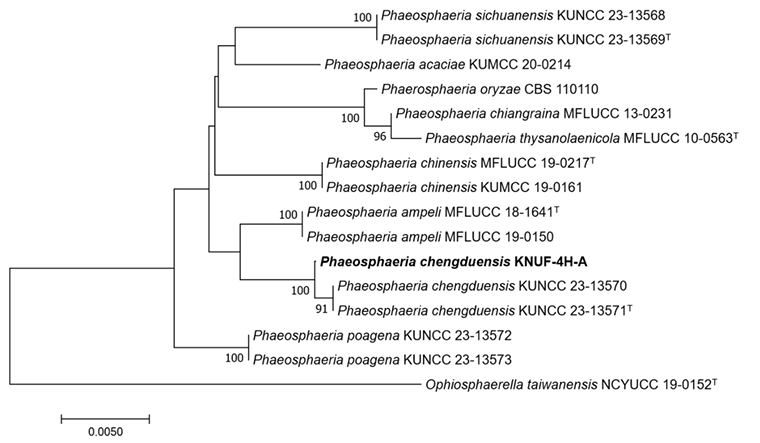
The obtained sequence of the ITS regions was 558 bp, exhibiting 100% and 97.2% similarities with those of P. chengduensis KUNCC 23-13570T and P. poagena CBS 136771, respectively. The SSU gene (993 bp) exhibited 100% similarity with P. musae Sawada LSU 1147, P. chinensis K.K. Zhang, S. Hongsanan, Tennakoon & N. Xie MFLUCC 19-0217, P. oryzae I. Miyake CBS 110110, P. chengduensis OR20640 and P. chengduensis OR206401. For the LSU gene (890 bp), the isolate exhibited 99.8-99.9% similarity with P. sinensis Jayasiri, E.B.G. Jones & K.D. Hyde MFLUCC 18-1552, P. chiangraina Phook. & K.D. Hyde MFLUCC 13-0231, P. thysanolaenicola Phook. & K.D. Hyde MFLUCC 10-0563, and P. chengduensis KUNCC 23-13570T. For the RPB2 gene, the obtained sequence was 829 bp and exhibited 97.9% similarity with P. chengduensis KUNCC 23-13570T. Regarding the TEF1-α gene, the obtained sequence was 858 bp and exhibited 100%, 97.6%, and 97.0% similarities with P. chengduensis KUNCC 23-13570T, P. cycadis Wanas., Phookamsak & K.D. Hyde KUMCC 18-0161, and P. chinensis MFLUCC 18-1552, respectively. These results demonstrated that none of the five gene sequences alone allowed for the precise identification of strain KNUF-4H-A at the species level. Recently, combined sequences of the ITS region, and the SSU, LSU, TEF1-α, and RPB2 genes were successfully used to classify several new members of the family Phaeosphaeriaceae [16]. The same approach was applied in our study for phylogenetic analysis. In the constructed maximum likelihood (ML) phylogenetic tree, a monophyletic clade composed of the KNUF4H-A isolate and two P. chengduensis strains (KUNCC 23-13571T and KUNCC 23-13570) with a high bootstrap value of 99%–100% unequivocally indicated that they belong to the same species (Fig. 2). The sequences of RPB2 or TEF1-α and RPB2 genes were not available for the closely related Phaeospaeria species, namely P. ampeli Tennakoon, C.H. Kuo & K.D. Hyde and P. chinensis, as the close neighbors of KNUF-4H-A. Therefore, an additional ML phylogenetic tree was constructed using the ITS, LSU, and SSU sequences. Similar to its position in the above-described tree, strain KNUF-4H-A clustered with the two strains of P. cheugduensis (Fig. 3). Collectively, the results of our molecular analyses demonstrated that the isolate belongs to P. cheugduensis.
Fig. 2
Maximum-likelihood phylogenetic tree of strain KNUF-4H-A based on the concatenated sequences of internal transcribed spacer (ITS) regions, and large subunit rDNA (LSU), small subunit rDNA (SSU), translation elongation factor 1-α (TEF1-α), and RNA polymerase II second largest subunit (RPB2) genes, showing the phylogenetic position of the new isolate among Phaeosphaeria species. The numbers above the branches represent the bootstrap values (>70%) obtained for 1,000 replicates. The isolated strain is indicated in bold. Ophiosphaerella taiwanensis SICAUCC 23-0003 was used as an outgroup. Bar, 0.01 substitutions per nucleotide position. ʻT’indicates type strain.
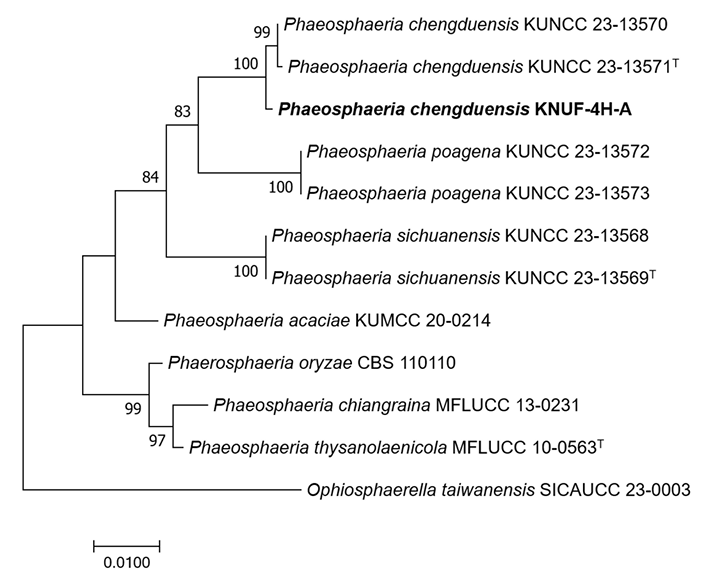
Fig. 3
Maximum-likelihood phylogenetic tree of strain KNUF-4H-A based on the concatenated sequences of internal transcribed spacer (ITS) regions, as well as the large subunit rDNA (LSU) and small subunit rDNA (SSU) genes, showing the phylogenetic position of the new isolate among Phaeosphaeria species. The numbers above the branches represent the bootstrap values (>70%) obtained for 1000 replicates. The isolated strain is shown in bold. Ophiosphaerella taiwanensis NCYUCC 190152T was used as an outgroup. Bar, 0.005 substitutions per nucleotide position. ʻT’indicates type strain.
The members of the genus Phaeosphaeria are known plant pathogens responsible for inducing various diseases across a wide range of hosts and are commonly found in decaying plant material, deadwood, or organic debris from monocotyledons [16]. Previous studies have identified P. chengduensis as a saprobe, isolated from dead twigs of an unidentified deciduous host. P. poagena is also saprobic, typically found on deceased bamboo, whereas P. sichuanensis was identified on dead leaves of Pandanaceae [16]. In Korea, three species of Penicillium were identified in Korean Muljara (Muljarus japonicas Vuillefroy), European oil beetle (Meloe proscarabaeus Linnaeus), and weevil (Lixus imperessiventris Roelofs) [30], along with three new species of Mucor found in crickets (Gryllus sp.) [31]. Recently, Monochaetia mediana S.Y. Lee & H.Y. Jung was isolated from the hairy long-horned toad beetle (Moechotypa diphysis Pascoe) in Korea [22]. However, to the best of our knowledge, no previous studies had reported on the occurrence of fungal species on the citrus flower chafer (Gametis jucunda). Although members of the genus Phaeosphaeria have been identified in various habitats worldwide [16], the identified strain KNUF-4H-A was isolated from the citrus flower chafer (Gametis jucunda) in Korea. Additionally, sexual morphological characteristics were not observed, but cultural and molecular analyses enabled the identification of strain KNUF-4H-A as P. chengduensis. In conclusion, our findings offer new perspectives into the taxonomic diversity within the genus Phaeospaeria in Korea and provide new insights into the previously undocumented asexual morphology of P. chengduensis KUNCC 23-13571T. Nevertheless, additional morphological analyses at the species level are required to further validate our findings. Moreover, further research is required to enhance our understanding of the interactions between insects and fungi, along with their classification, etiology, ecology, pathogenicity, and potential activities. To the best of our knowledge, our study constitutes the first record of Phaeosphaeria chengduensis KNUF-4H-A in Korea.