INTRODUCTION
Lindera obtusiloba Blume is a shrub widely distributed in the understory vegetation of deciduous forests in East Asia, including Korea [1]. This species contains compounds with pharmacological potential, including antioxidant, anti-allergic, and anti-inflammatory properties [2]. Endophytic fungi are a highly diverse polyphyletic group of fungi that inhabit plant tissues without causing any apparent symptoms [3]. These fungi have gained significant attention as metabolites synthesized by these include plant-derived metabolites with anticancer, antibiotic, and antimicrobial activities [4]. One such antioxidant compound, salidroside, extracted from L. obtusiloba, has been found to be produced by the endophytic fungus Phialocephala fortinii isolated from plants of the genus Rhodiola [5,6].
In Korea, 12 species of endophytic fungi from 11 genera have been reported from the leaves of L. obtusiloba [7]. However, further studies are required to explore the diversity of endophytic fungi in other regions and tissues of this plant. This study describes the molecular phylogenetic analysis and morphological characteristics of two previously unrecorded endophytic fungi in Korea, Colletotrichum citricola and Valsa ceratophora, isolated from the leaves and twigs of L. obtusiloba.
MATERIALS AND METHODS
Sample collection and preparation. Leaves and twigs of L. obtusiloba Blume were collected from two locations in Korea: Samcheok-si, Gangwon-do, 37°13'19.3"N 129°00'35.8"E in March 2023, and Jecheonsi, Chungcheongbuk-do, 36°54'02.4"N 128°05'37.0"E in July 2023. Healthy leaf and twig samples were collected without any symptoms or damage. The samples were placed in polyethylene bags and transported to the laboratory within 12 hours.
Surface sterilization and fungal isolation. The leaf samples were washed with tap water to remove surface contaminants. The surface sterilization process involved immersion in 35% H2 O2 solution for 40 seconds for leaves and 90 seconds for twigs, followed by immersion in 70% ethanol for 30 seconds [8]. After surface sterilization, the leaf samples were cut into 1.5 cm pieces and placed on potato dextrose agar (PDA; Difco Lab., Detroit, USA). These samples were incubated at 25℃ in the dark to induce endophytic fungal growth.
Culturing and morphological characterization. Mycelia growing from inside the leaf and twig pieces were sub-cultured to obtain pure fungal strains. These pure strains were grown on both malt extract agar (MEA; Kisan bio, Seoul, Korea) and PDA to observe the morphological characteristics of the colonies at 25℃ in the dark. Colony morphology was observed visually, and fungal spore-forming structures were examined using a light microscope (Axio Imager A2, Carl ZWISS, Oberkochen, Germany).
DNA extraction and molecular identification. For molecular phylogenetic identification, DNA was extracted from mycelia using a HiGene Genomic DNA Prep Kit (Biofact, Daejeon, Korea). Specific DNA regions necessary for strain identification were amplified by PCR using the specific primer sets as follows: internal transcribed spacer (ITS) with ITS1F/ITS4 [9,10], β-tubulin (TUB2) with Bt2a/Bt2b [11], chitin synthase (CHS) with CHS-79F/CHS-345R [12], and glyceraldehyde-3-phosphate dehydrogenase (GAPDH) with GDF/GDR [13]. The PCR conditions were 95℃ for 2 minutes, followed by denaturation at 95℃ for 20 seconds, annealing at temperatures specific to each primer set (50℃ for ITS, 55℃ for TUB2, 57℃ for CHS, and 58℃ for GAPDH) for 40 seconds, extension at 72℃ for 1 minute, and a final extension at 72℃ for 5 minutes. The PCR products were electrophoresed on a 1.5% agarose gel to verify the amplification and fragment size.
DNA sequencing and phylogenetic analysis. DNA sequencing was performed by Solgent Co., Ltd. (Daejeon, Korea). The obtained sequences were compared with those in the National Center for Biotechnology Information (NCBI) database using the Basic Local Alignment Search Tool (BLAST). Phylogenetic trees were constructed using the neighbor-joining method in MEGA11 software [14]. The isolated strains were deposited at the National Institute of Biological Resources (NIBR), and the sequences were registered with NCBI.
RESULTS AND DISCUSSION
Colletotrichum citricola F. Huang, L. Cai, K.D. Hyde & Hong Y. Li, Fungal Diversity 61: 67 (2013) [MB#803018] Fig. 1
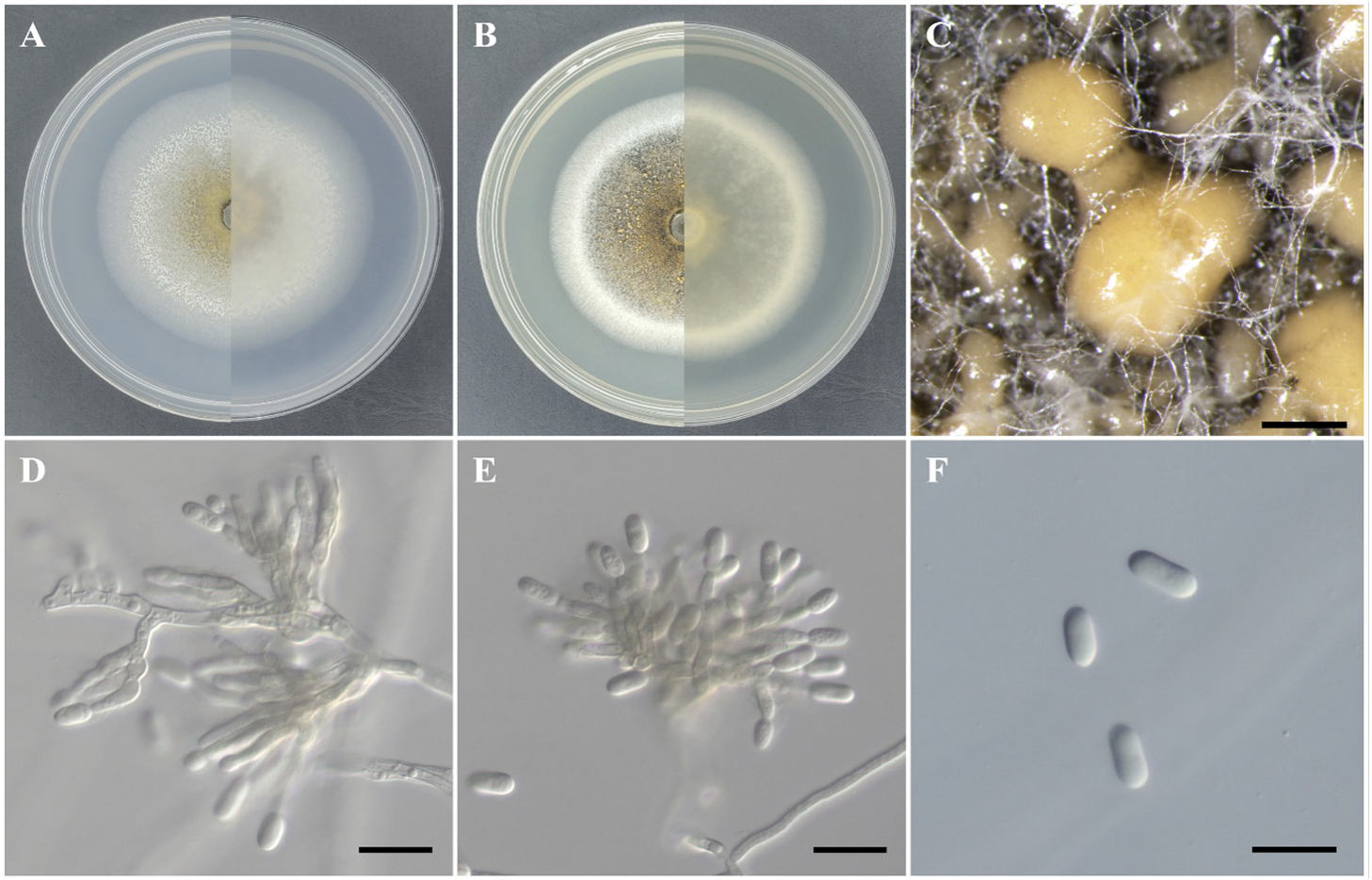
Morphological characteristics of strain KNUE23P615: After 7 days of incubation at 25℃ in the dark, colonies on MEA reached a diameter of 46.0–48.0 mm. The colony surface was brown at the center, gradually fading towards the edges and surrounded by white filamentous mycelia. The reverse side of the colony was dark brown at the center and white at the edges. The colonies were flat with entire margins (Fig. 1A). On PDA, the colony diameter ranged from 34.2-38.0 mm. The colony surface was yellow at the center and surrounded by white filamentous mycelia and black spots. The reverse side of the colony was yellow at the center and white at the edges. The colonies were flat with entire margins (Fig. 1B). The conidiomata developed as orange or light brown, sticky, and slimy masses (Fig. 1C). The conidiophores were hyaline and variously branched, and conidiogenous cells formed transparently as the tips of the conidiophores tapered (Fig. 1D and 1E). The conidia were cylindrical with rounded ends, hyaline, and smooth-walled. The size of the spores was (9.8-) 12.0 (-14.5)×(4.9-) 6.1 (-7.4) μm (n=20) (Fig. 1F).
Fig. 1
Morphology of Colletotrichum citricola KNUE 23P615. A-B Colonies after 7 days of growth at 25℃. The left side shows the front view, and the right side shows the reverse view of malt extract agar (A) and potato dextrose agar (B). C: Conidiomata (Bar=500 μm), D-E: Conidiophores and conidiogenous cells (Bar=20 μm), F: Conidia (Bar=20 μm).
Specimen examined: Jecheon-si Chungcheongbuk-do, Korea, 36°54'02.4"N 128°05'37.0"E, July 28, 2023, C. citricola, isolated from the leaf of L. obtusiloba, strain KNUE23P615, NIBRFGC000510709, GenBank No. PP795380 (ITS), PP801054 (TUB2), PP801055 (CHS), and PP081056 (GAPDH).
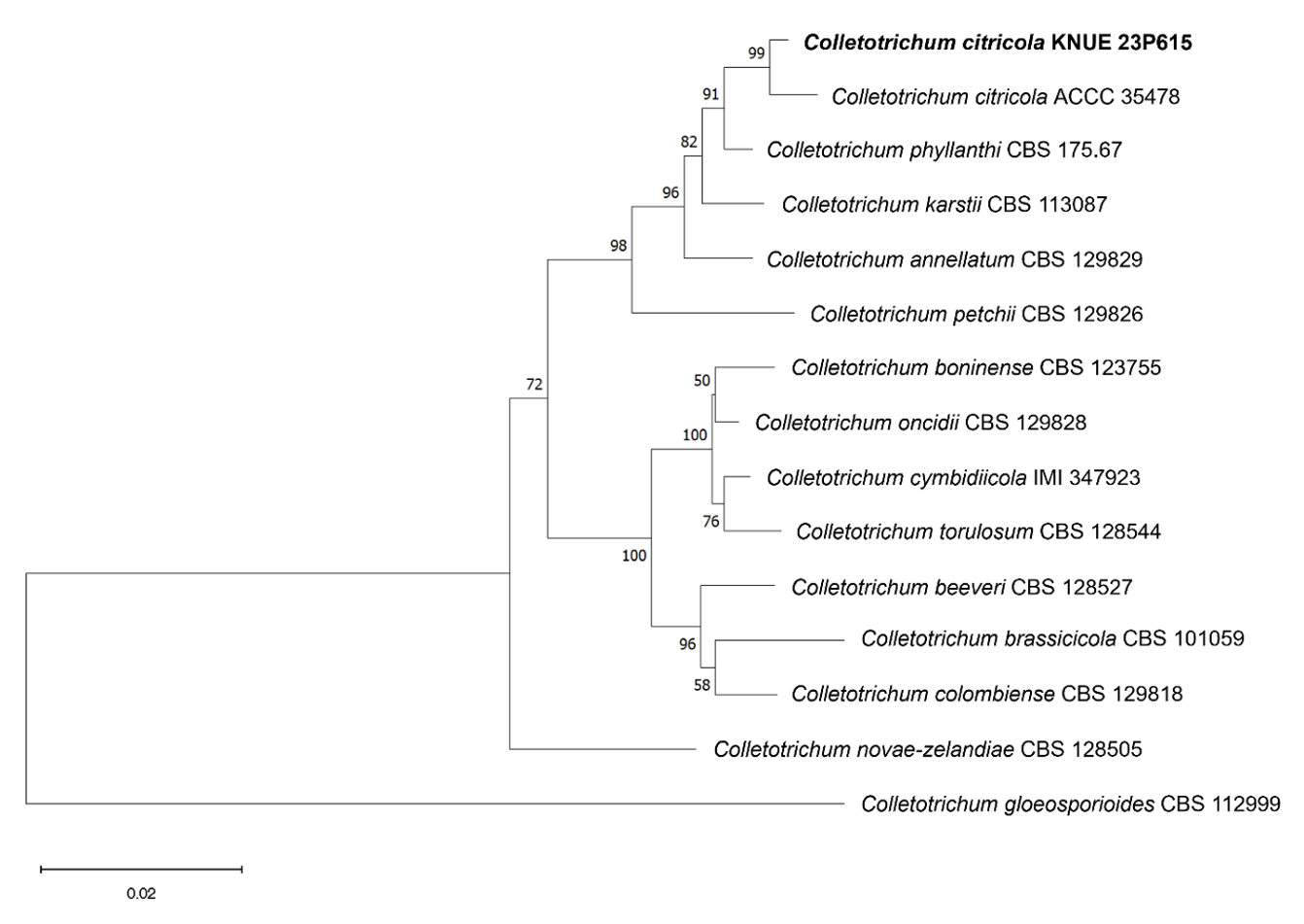
Notes: C. citricola was first reported in 2013 as an epiphyte isolated from the leaves of Citrus unshiu [15]. It has since been isolated from the genus Dendrobium in China and was recently identified as the cause of leaf spots in Cavendish bananas, demonstrating that its potential for pathogenicity depends on the host plant [16,17]. Strain KNUE23P615 exhibited a distinctly wider conidial width of approximately 6 μm compared to that of the closely related C. phyllanthi, consistent with the original description (Table 1). The ITS sequence of strain KNUE23P615 showed 99.65% identity with that of C. citricola A12 (MT269527), and the TUB2 sequence showed 99.09% identity with that of C. citricola FS1-3 (OL361858). The CHS sequence showed 100% identity with that of C. citricola FS1-3 (OL361855), and the GAPDH sequence showed 99.49% identity with that of C. citricola PAFQ13 (MG747980). In a neighbor-joining phylogenetic tree constructed using the combined ITS, TUB2, CHS, and GAPDH sequences, KNUE23P615 formed a clade with C. citricola ACCC 35478 (Fig. 2).
Table 1
Morphological characteristics of strains KNUE 23P615 and Colletotrichum citricola CBS 134228
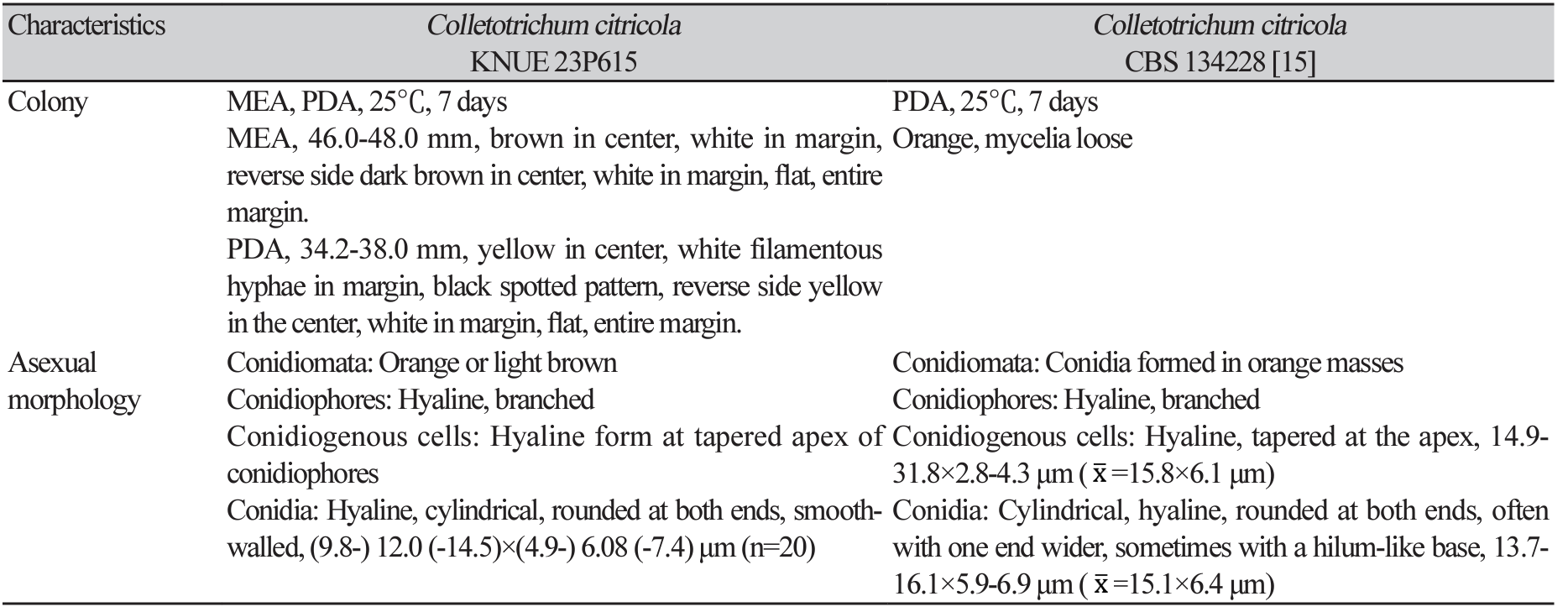
Fig. 2
Neighbor-joining phylogenetic tree of Colletotrichum citricola KNUE 23P615. The tree is based on concatenated sequences of the internal transcribed spacer (ITS), β-tubulin (TUB2), chitin synthase (CHS), glyceraldehyde-3-phosphate dehydrogenase (GAPDH) DNA sequences. Colletotrichum gloeosporioides was used as an outgroup. The numbers on the nodes represent bootstrap values greater than 50% (1,000 replicates).
Valsa ceratophora Tul. & C. Tul., Selecta Fungorum Carpologia, Tomus Secundus. Xylariei - Valsei - Sphaeriei 2: 191 (1863) [MB#141122] Fig. 3
Morphological characteristics of strain KNUE23N362: After 7 days of growth at 25℃ in the dark, colonies on MEA reached a diameter of approximately 90 mm, with very faint mycelia at the edges but covering most of the medium. The colony surface was light greyish-yellow at the center, lighter towards the edges, and showed alternating light and dark bands. The reverse side of the colony was dark chestnut brown in the center, becoming lighter and ivory-colored towards the edges. The colonies were flat with a smooth surface (Fig. 3A). On PDA, the colony diameter was approximately 90 mm, with low mycelium density at the edges but covering almost the entire medium. The colony surface was light yellowish-brown at the center and filamentous; light ivory mycelia were observed at the edges. The reverse side of the colony became lighter from the center to the edges. The colonies were flat with a rough surface (Fig. 3B). The conidiomata were scattered, irregularly shaped, and protruded from the colony. They were black and hard and sometimes had yellowish-brown mucilage on their sides (Fig. 3C). The conidia were hyaline, long, cylindrical with rounded ends, smooth-walled, and sometimes slightly curved. The size of the spores was (3.73-) 4.04 (-4.98)×(0.81-) 1.31 (-1.56) μm (n=20) (Fig. 3D).
Specimen examined: Samcheok-si, Gangwon-do, Korea, 37°13’19.3”N 129°00’35.8”E, March 31, 2023, Valsa ceratophora, isolated from a twig of Lindera obtusiloba strain KNUE23N362, NIBRFGC000510698, GenBank No. PP795379 (ITS), and PP795381 (LSU).
Notes: The genus Valsa is a teleomorph of the genus Cytospora, with most species reported between the mid-1800s and mid-1900s. V. ceratophora was first reported in 1863 from dead oak bark in France, and its anamorph, C. ceratophora, was first reported in 1879 [18,19]. V. ceratophora has been reported in various countries worldwide, inhabits a wide range of plants, and is opportunistic, causing canker diseases [20]. Although no records exist of the culture and conidial morphology of V. ceratophora on media, its cylindrical and curved conidia are similar to those of the new Cytospora species reported in 2018 and 2019 [21,22]. The high sequence similarity of ITS and LSU further supports the identification of KNUE23N362 as V. ceratophora. The ITS sequence showed 99.83% identity with that of V. ceratophora CBS 109777 (MH862834), and the LSU sequence showed 100% identity with that of V. ceratophora CBS 397.36 (MH867343). In the neighbor-joining phylogenetic tree constructed using the combined ITS and LSU sequences, KNUE23N362 formed a group with V. ceratophora CBS 397.36 (Fig. 4).
Fig. 3
Morphology of Valsa ceratophora KNUE 23N362. A-B Colonies after 7 days of growth at 25℃. The left side shows the front view and the right side shows the reverse side of malt extract agar (A) and potato dextrose agar (B). C: Conidiomata (Bar=500 μm), D: Conidia (Bar=2 μm).
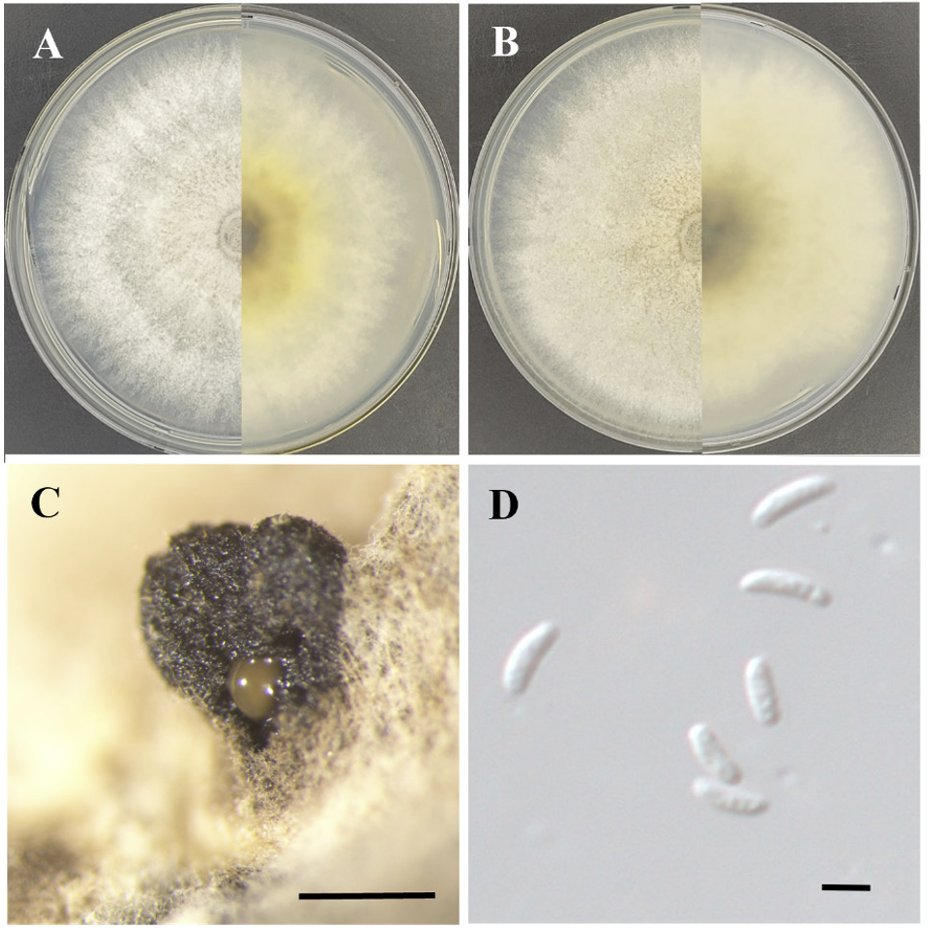
Fig. 4
Neighbor-joining phylogenetic tree of Valsa ceratophora KNUE 23N362. The tree is based on concatenated sequences of the internal transcribed spacer (ITS) and large subunit rDNA (LSU) sequences. Diaporthe vaccinii was used as an outgroup. The numbers on the nodes represent bootstrap values greater than 50% (1,000 replicates).

In this study, we characterized two previously unrecorded endophytic fungi, C. citricola, and V. ceratophora, isolated from L. obtusiloba in Korea. Morphological observations, combined with molecular phylogenetic analysis using ITS, TUB2, CHS, GAPDH, and LSU sequences, confirmed the identity of these fungi. Colletotrichum species are known pathogens; however, novel Colletotrichum species have also been reported as endophytes [23,24]. Some Colletotrichum species have been identified as both pathogens and endophytes [25]. Similarly, Valsa species have been reported to cause Valsa cankers on fruit trees [26,27]. However, several species, including Valsa friesii and Cytospora chiangmaiensis, have been identified as endophytes with antagonistic potential against plant pathogenic fungi [28,29]. The discovery of C. citricola and V. ceratophora as endophytes expands our understanding of the relationship between these fungal species and host plants. It also provides information on the diversity of fungal species associated with L. obtusiloba and enhances the knowledge of fungal biodiversity in Korea. However, further studies are needed to explore the ecological roles of these fungi and their potential applications.

