Endophytic fungi form symbiotic relationships within plant tissues, causing no negative symptoms or diseases [1]. Approximately 84% of endophytic fungi belong to the phylum Ascomycota and 10% belong to the phylum Basidiomycota [2]. Although the specific mechanisms by which these fungi confer benefits OPEN ACCESS pISSN : 0253-651X eISSN : 2383-5249 Kor. J. Mycol. 2024 March, 52(1): 13-18 https://doi.org/10.4489/kjm.520102 Received: February 07, 2024 Revised: March 22, 2024 Accepted: March 24, 2024 © 2024 THE KOREAN SOCIETY OF MYCOLOGY. This is an Open Access article distributed under the terms of the Creative Commons Attribution Non-Commercial License (http: //creativecommons.org/licenses/by-nc/4.0/) which permits unrestricted non-commercial use, distribution, and reproduction in any medium, provided the original work is properly cited. to plants remain largely unknown, their documented roles include providing protection against pathogens and herbivores and improving resistance to abiotic stresses, such as drought, high salinity, and temperature variations. This highlights their potential application in sustainable agriculture [3]. The diverse secondary metabolites and enzymes produced by endophytic fungi have high industrial value, particularly in the field of natural pharmaceutical development and discovery [4-6]. Despite their importance, the diversity of endophytic fungi remains underexplored in many regions. This knowledge gap hinders our understanding of the full spectrum of fungal diversity and their potential applications. Therefore, studying previously unrecorded endophytic fungal species is vital to expand our understanding of their roles in ecosystems and their potential use in various industries.
In Korea, studies on endophytic fungi are becoming increasingly comprehensive, with an expanding focus on their taxonomy and ecological interactions [7]. This study aimed to describe the morphological features and perform a phylogenetic analysis of the previously unrecorded endophytic fungus Aequabiliella effusa in Korea, which was discovered during the isolation of endophytes from Larix gmelinii var. olgensis.
In June 2023, the twigs of L. gmelinii var. olgensis were obtained from Danyang, Chungcheongbuk-do, Korea. Healthy plant tissue samples without any symptoms of disease were collected and transported to the laboratory within 24 h in polyethylene bags for fungal isolation.
The samples were washed with distilled water to remove debris and subjected to surface sterilization with 30% H2 O2 for 90 s. A secondary surface sterilization was performed using 70% ethanol for 30 s. After sterilization, the samples were cut into 1.5 cm pieces and placed on potato dextrose agar (PDA; Difco Lab., Detroit, USA) for cultivation. Following the observation of fungal growth in the interior of the plant samples, subculturing was conducted to isolate pure fungal strains.
For the observation of colony morphology and fungal microstructures, the fungal cultures were incubated on potato dextrose agar and malt extract agar (MEA; Kisan bio, Seoul, Korea) at 25℃ in the dark for 7 days. The microstructures were observed under a light microscope (Axio Imager A2, Carl ZWISS, Oberkochen, Germany).
DNA was extracted from the mycelia using the HiGene Genomic DNA Prep Kit (BioFACT, Daejeon, Korea) according to the manufacturer’s instructions. For phylogenetic analysis, two DNA regions were amplified using Polymerase Chain Reaction (PCR): the internal transcribed spacer (ITS) using ITS1F/ITS4 primers [8,9] and the large subunit ribosomal DNA (LSU) region using LROR/LR16 primers [10].
The PCR conditions for the ITS and LSU regions started with an initial DNA denaturation at 95℃ for 2 minutes, followed by 35 cycles of 95℃ for 20 seconds, 50℃ (for ITS) or 44℃ (for LSU) for 40 seconds, and 72℃ for 1 minutes, with a final DNA extension step at 72℃ for 5 minutes.
The PCR products were electrophoresed on 1.5% agarose gels for 20 min to verify DNA amplification by assessing the size of the DNA fragments. Sanger sequencing was performed by SolGent Co., Ltd. (Daejeon, Korea). The DNA sequences of the isolates were compared with those in the GenBank database of the National Center for Biotechnology Information (NCBI) using the Basic Local Alignment Search Tool (BLAST). The DNA sequences of each isolate were concatenated, and a neighbor-joining phylogenetic tree was constructed using MEGA7 software with 1,000 bootstrap replications [11]. The strains were deposited at the National Institute of Biological Resources (NIBR) and the DNA sequences were registered in the NCBI database.
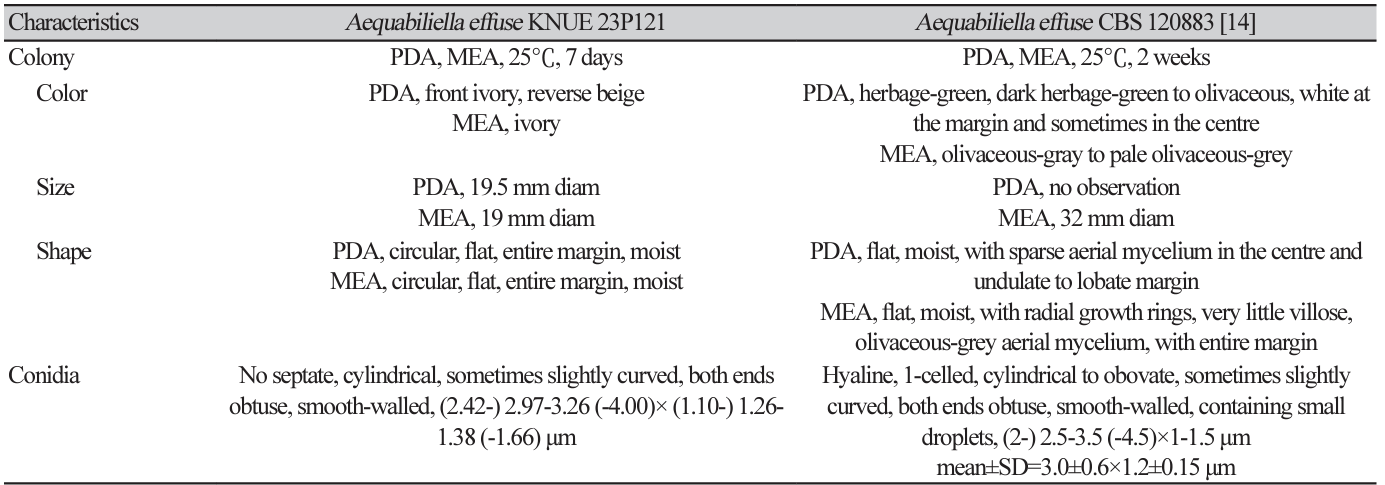
Phylogenetic analysis was performed using DNA sequences from the ITS and LSU regions. The ITS sequence of strain KNUE 23P121 showed 99.44% similarity with A. effusa CBS 120883 (NR_132005), and the LSU sequence was 99.4% identical to A. effusa CBS 120883 (NR_056966). Strain KNUE 23P121 formed a cluster with A. effusa CBS 120883 in a neighbor-joining phylogenetic tree constructed from the combined ITS and LSU sequences (Fig. 1).
Fig. 1
Neighbor-joining phylogenetic tree of Aequabiliella effusa KNUE 23P121 based on concatenated sequences of internal transcribed spacer (ITS) and large subunit ribosomal DNA (LSU) regions. Phytophthora moyootj was used as an outgroup. The numbers on the nodes represent bootstrap values greater than 50% (1,000 replicates).
Aequabiliella effusa (Damm & Crous) Crous, Persoonia 34: 225 (2015) [MB#812476]
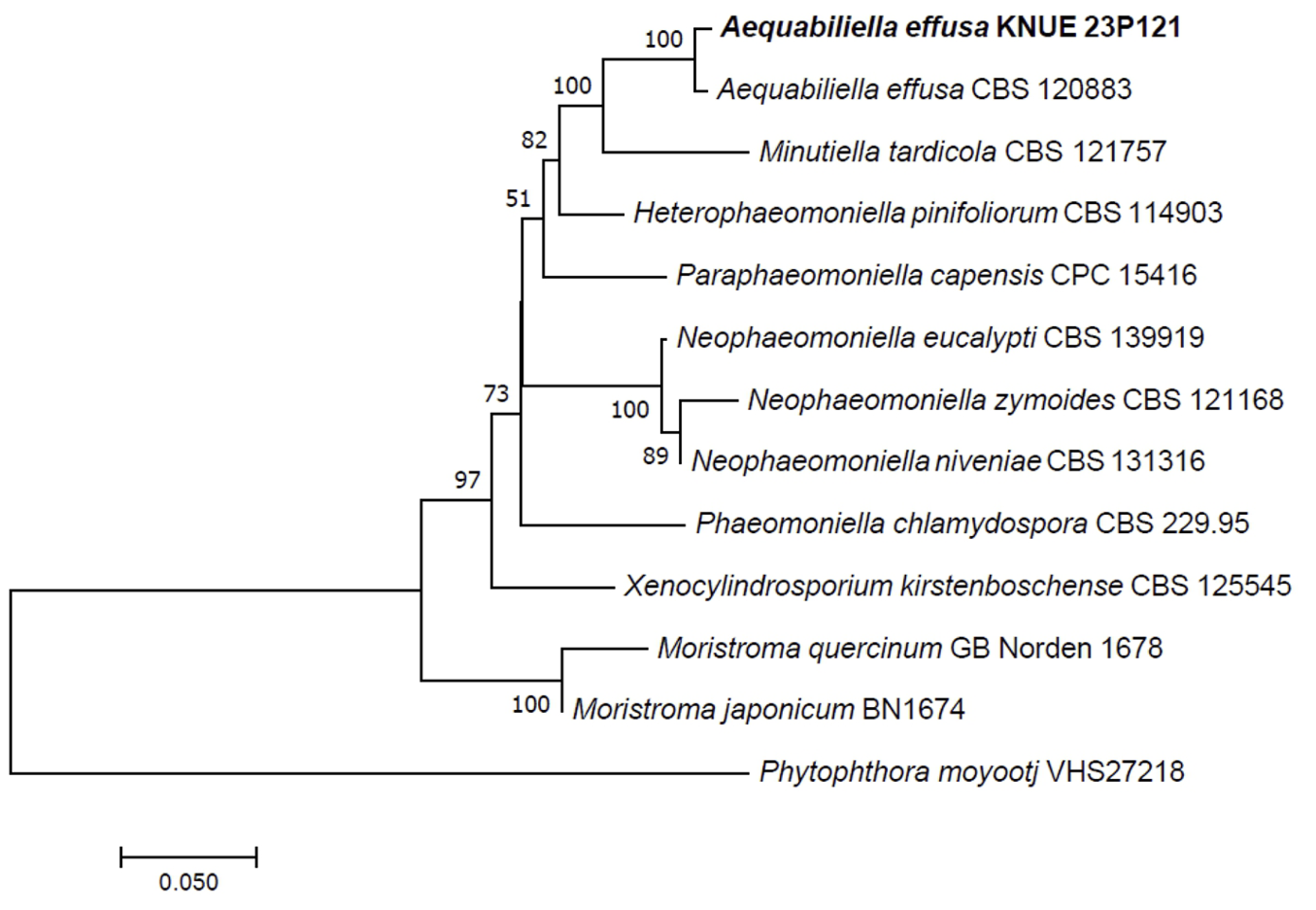
On PDA, the colony morphology appeared circular, with flat elevation and entire margin, displaying moist and mucilaginous texture. The colony diameter was 19.5 mm, with an ivory color (Hex code: E1E4D1) and beige color (Hex code: E3E2C6) on the reverse side (Fig. 2A). On MEA, the colony morphology remained circular with flat elevation, entire margin and moist texture. The colony diameter was 19 mm, with both the surface and reverse sides presenting an ivory color (Hex code: DEE4D7) (Fig. 2B). The mycelia were hyaline with septate and smooth cell walls. The conidia were cylindrical, with both ends rounded and slightly curved. The conidial walls were smooth without septa. The dimensions of the conidia ranged from (2.42-) 2.97-3.26 (-4.00)×(1.10-) 1.26-1.38 (-1.66) μm (Fig. 2C and D).
Fig. 2
Morphology of Aequabiliella effusa KNUE 23P121. A: A colony on potato dextrose agar (PDA), left side is front side and right side is reverse side after 7 days at 25℃, B: A colony on malt extract agar (MEA), left side is front side and right side is reverse side after 7 days at 25℃, C, D: Conidia. Scale bars: 10 μm.
Specimen examined: Danyang, Chungcheongbuk-do, Korea, 37°00'02.9"N 128°16'15.6"E, June 2, 2023, Aequabiliella effusa, isolated from twig of Larix gmelinii var. olgensis, KNUE 23P121, and NIBRFGC000510708; GenBank No. PP217727 (ITS) and PP196669 (LSU).
Notes: Aequabilella effusa, initially reported as Phaeomoniella effusa in 2010, was reclassified in 2015 [12,13]. The first isolation was from the necrotic parts of peach trees and was later reported in branches of Pinus sylvestris in Spain [12,14]. The colonies of KNUE 23P121 showed slight differences in color compared to the original description; KNUE 23P121 was ivory and beige, whereas CBS 120883 was herbage-green and olivaceous. However, the shape and size of the conidia are consistent with those in the original description (Table 1).
In the present study, endophytic fungal strains isolated from the twigs of L. gmelinii var. olgensis were isolated and identified based on their morphological and molecular characteristics. The strain KNUE 23P121 was identified as A. effusa. To the best of our knowledge, these fungal species were not reported previously in Korea. In this study, we aimed to expand our knowledge of fungal diversity in Korea.