INTRODUCTION
The class Sordariomycetes in the phylum Ascomycota, comprises highly diversified fungal groups that play critical roles in the ecosystem [1]. It is well known that the Sordariomycetes is the second largest class of the phylum Ascomycota [2]. Among the Ascomycota classes, Sordariomycetes exhibits a high substitution rate resulting from an acceleration of the speciation process over time, as compared to Dothideomycetes and Leotiomycetes [3]. Sordariomycetes contains 1,331 genera distributed across 105 families, 32 orders and six subclasses [4]. The subclasses include Diaporthomycetidae, Hypocreomycetidae, Lulwor-thiomycetidae, Meliolomycetidae, Sordariomycetidae and Xylariomycetidae. Members of the Sordariomycetes are ubiquitous and cosmopolitan, which function in almost all ecosystems as pathogens and endophytes of plants, arthropods and mammals, as mycoparasites and as saprobes involved in decomposition and nutrient cycling [5, 6].
The objectives of this study were to analyze the unidentified fungi included in the Sordariomycetes class as well as to determine the phylogenetic placement and illustration of these distinctive fungi. To this end, we isolated numerous morphologically distinct fungal strains during an investigation of unrecorded fungal species in Korea. The results of this screening of unrecorded fungal strain would not only be a useful tool for the identification of indoor Sordariomycetes but also provide a new insight into phylogeny of the class Sordariomycetes. To the best of our knowledge, three soil-inhabiting fungi identified through this study are new to Korea.
MATERIALS AND METHODS
Soil sample collection and fungal isolation
The soil samples were collected from Daegu, Korea in 2017. The soils were collected from depth (15 to 30 cm) using sterile spatula and transferred to polythene zipper bags and then brought to the laboratory and stored under refrigeration. For analysis, the soils were suspended in 10 mL of sterile distilled water and gently vortexed. Then, the suspension was serially diluted and spread on potato dextrose agar (PDA; Difco, Detroit, MI, USA) plates. The PDA plates were incubated 3 days at 25°C temperature after which the single fungal colonies were transferred to new plates and exposed to the same incubation temperature. Of these isolates, BH-06, 17-039 and BE12-1 were selected for further morphological and molecular phylogenetic analyses.
Morphological characterization
The morphological characteristics were studied on PDA, oatmeal agar (OA), malt extract agar (MEA) and synthetic nutrient agar (SNA) for 7 days at 25°C. All the media were used to study the morphology of the isolate BE12-1. But in case of isolates BH-06 and 17-039 they were cultured on PDA media for 7 days at 25°C. After the incubation, the diameters of the colonies were measured and colony color was noted for each of the isolates. A light microscope was used to observe the mycological characteristics of the two isolates.
Genomic DNA extraction, PCR amplification and sequencing
The three genera of Sordariomycetes were subjected to molecular identification based on multiple genes. Genomic DNA was extracted from three isolates namely, BH-06, 17-039 and BE12-1 by using the HiGene Genomic DNA prep kit (BIOFACT, Daejeon, Korea) as per the manufacturer’s instructions. The isolate BE12-1 were amplified for four gene markers with the primers RPB2AM-1bf and RPB2AM-7R [7] for the second largest subunit of DNA-directed RNA polymerase II (RPB2) gene region, ITS5 and ITS4 [8] for the internal transcribed spacer regions (ITS) and intervening 5.8S nrRNA gene region, NL1 and NL4 [9] for the D1/D2 domains of the 28S nrDNA large subunit (LSU), and T1 [10] and TUB4Rd [11] for the partial β-Tubulin (TUB2) gene region. The PCR conditions were the same as those described by Wang et al. [12]. In case of isolate BH-06 was amplified using primers ITS1F and ITS4, while isolate 17-039 was amplified by using the LR3R and LR5 for nrLSU [13-15] and nrSSU, NS1 and NS8 for nrSSU [7]. The amplified PCR product was purified with EXOSAP-IT (Thermo Fisher Scientific, Waltham, MA, USA) and sequenced by SolGent (Daejeon, Korea) using an ABI 3730XL DNA analyzer (Applied Biosystems, Foster City, CA, USA). The sequences obtained from BE12-1, BH-06 and 17-039 were deposited in NCBI GenBank.
Phylogenetic analyses
The sequences of the Korean our isolates were compared with reference sequences from the GenBank database of the National Center for Biotechnology Information (NCBI), using the basic local alignment search tool (BLAST). The evolutionary distance matrices were generated based on the Kimura’s neighbor-joining algorithm model [16]. The phylogenetic analyses were performed using the program MEGA 7 [17] with bootstrap values based on 1,000 replications.
RESULTS AND DISCUSSION
Chaunopycnis alba Gams W. Persoonia 11:75–79 (1980)
Morphology of the isolate BH-06
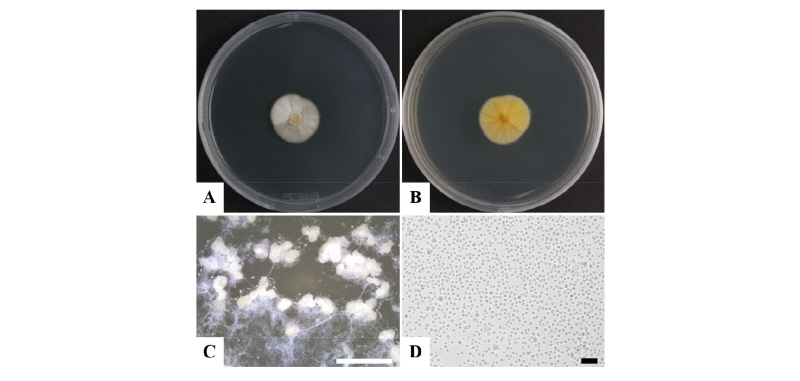
Concerning our strain isolated from soil, we observed the mycelial colonies to be white, yellowish and smooth, with unicellular conidia on PDA. They also produced numerous white and globular pycnidia when the PDA plates were incubated for 2~3 weeks (Fig. 1). The pycnidia (n = 20) were white in color and globular in structure with a diameter of 40~ 280 µm. The conidia were unicellular, hyaline and globose. The conidiophores produced conidia holoblastically (Fig. 1C, 1D). Previous results showed that Chaunopycnis alba produced colonies that were white and thinly floccose, with conidiomata embedded in the hyaline aerial mycelium, of irregular roundish shape, and featuring indistinct conidiophores with cylindrical, distally tapering phialides and one-celled hyaline conidia aggregated in slimy heads [18]. In contrast, colonies of our strain (BH-06) were white to pale yellow, floccose structured with hyaline mycelium, of round and wavy shape and featuring one-celled hyaline conidia (n = 30), 1.20~2.33 µm in diameter (Fig. 1). The morphology of the isolate BH-06 was similar to that of Chaunopycnis alba (Table 1).
Molecular phylogeny of isolate BH-06
Nucleotide sequences of ITS were analyzed to infer the evolutionary relationships of the isolates of BH-06 with the previously described species of Chaunopycnis alba (Table 2). The phylogenetic analysis (Fig. 2) clustered BH-06 closely with the previous Chaunopycnis alba which was also amplified based on the ITS (ITS1F/ITS4) ribosomal DNA sequence [18, 19]. The partial ITS sequence of the BH-06 strain consisted of 558 base pairs. The GenBank database also reflected a maximum 100% and 99% similarities with strains of Tolypocladium album (KU516596) and Chaunopycnis alba (JN628073). Hence, the results of phylogenetic analysis support the grouping of our isolate BH-06 with Chaunopycnis alba.
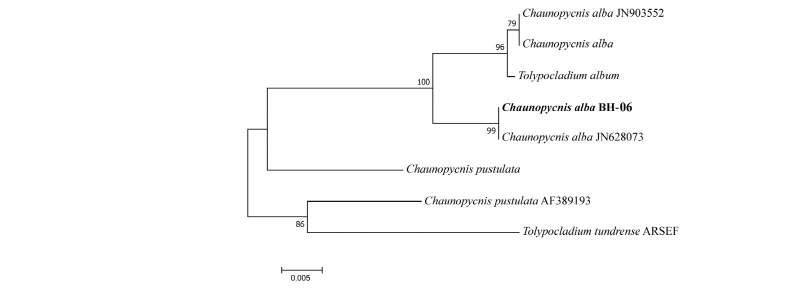
Fig. 2. Neighbor-joining phylogenetic tree based on internal transcribed spacer region sequences showing the relationships between Chaunopycnis alba and other members of the genus Chaunopycnis. The strain isolated in this study is indicated in bold. Bootstrap values are based on 1,000 replications. Bar, 0.005 substitutions per nucleotide position.
The identification of indole diterpenes in Chaunopycnis pustulata and Chaunopycnis alba extends the distribution of this metabolite class to a new phylogenetic lineage of the Clavicipitaceae. Chaunopycnis alba produces an indole diterpene which has potent neurotoxic effects in mammals [20]. To our knowledge, this is the first report of Chaunopycnis alba in Korea.
Myrothecium cinctum Estelle and Richard. Aerobiologia 13:227–234 (1997)
Morphology of the isolate 17-039
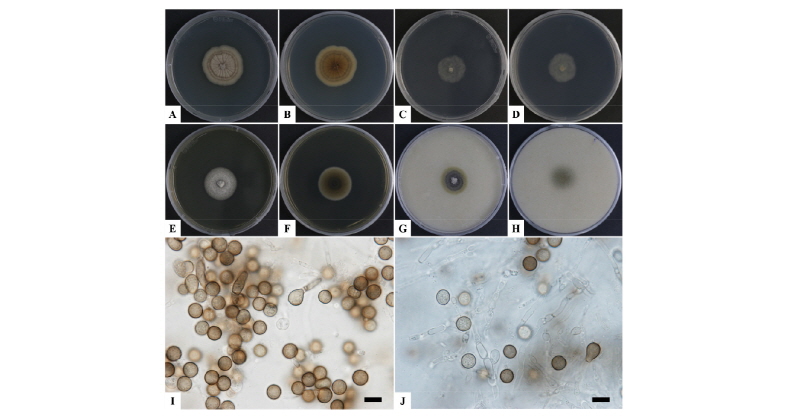
The isolate 17-039 exhibited white floccose mycelial colonies after 7 days culture on PDA at 25°C. The morphological characteristics were compared with those of previously described of Myrothecium cinctum [21]. Myrothecium cinctum, produces floccose and white mycelium with black sporodochia (cushion-shaped aggregations of conidiophores). The conidia start out pale yellow or green and develop deep green pigmentation as the spores matured. The conidiophores are born from the inner hyphal tissue of pycnidia, and produce conidia holoblastically. In contrast, our strain 17-039 produced white and floccose mycelial colonies of 38~44 mm diameter after 7 days culture on PDA (Fig. 3). The spores mature and develop green and deep green pigmentation with the black cushion shaped sporodochia. They also produce conidia holoblastically (Fig. 3D, 3E) and the spore (n = 30) size was 6.08~9.03 × 3.04~4.05 µm (Table 3).
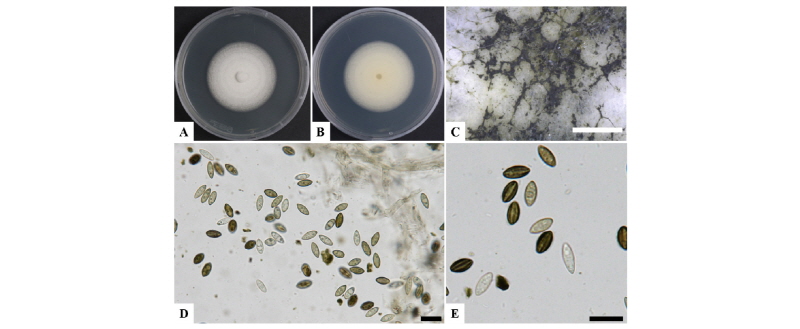
Fig. 3. Morphological characterization of Myrothecium cinctum (17-039) via a light microscope. A, Colony in front; B, Colony in reverse on potato dextrose agar (PDA); C, Colony appearance growing on PDA showing abundant sporodochia; D, E, Mycelia and spores on PDA (scale bars: C = 1,000 µm, D, E = 10 µm).
Molecular phylogeny of the isolate 17-039
The evolutionary relationship were inferred based on the large subunit (LSU) and small subunit (SSU) sequences of the isolate 17-039 in comparison with strains of the previously described species of Myrothecium cinctum (Table 2). Phylogenetic analyses (Fig. 4) suggested that the isolate is closely related to Myrothecium cinctum, but also a comparison with the reference sequences in NCBI GenBank revealed no nucleotide difference with M. cinctum strains (ATCC 22270, BBA 69182 for LSU and ATCC 22270 for SSU). Both results strongly support the grouping of our isolate with M. cinctum.
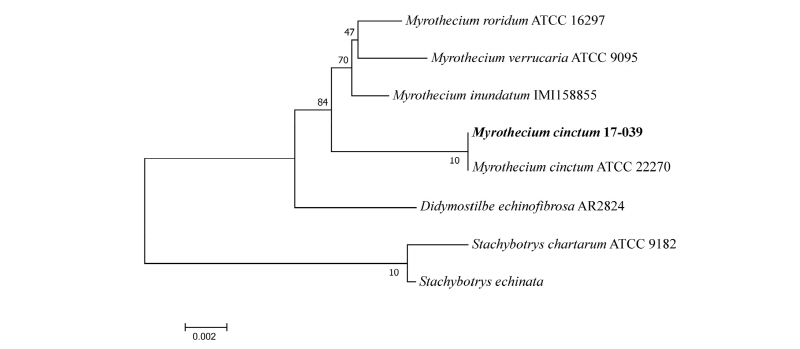
Fig. 4. Neighbor-joining phylogenetic tree based on small subunit and large subunit genes sequences showing the relationships between Myrothecium cinctum and members of the genus Myrothecium. The strain isolated in this study is indicated in bold. Bootstrap values are based on 1,000 replications. Bar, 0.002 substitutions per nucleotide position.
The Myrothecium are soil or leaf surface saprobes or weak plant pathogens causing leaf spot disease on various types of vegetation [22, 23]. The Myrothecium is also a prolific producer of bioactive secondary metabolites, with more than 20 reported compounds from M. roridum, and more than 30 compounds from M. verrucaria [24]. To our knowledge, this is the first report of M. cinctum in Korea.
Humicola olivacea Wang XW, Houbraken J, Groenewald JZ, Meijer M, Andersen B, Nielsen KF, Crous PW, Samson RA. Studies in Mycology 84:145–224 (2016)
Morphology of the isolate BE12-1
The isolate BE12-1 was observed with culturing the different media after 7 days incubation at 25°C. The morphological structures were compared with the strain of previously described (Table 4) species of Humicola olivacea [12]. The mycelium colony was greenish olivaceous; reverse black and greenish on OA; white and floccose mycelium on MEA; translucent mycelium on SNA; pale brown and yellow on PDA (Fig. 5). The average growth of fungal colonies were 36~62 mm in diameter in all the media. They produced plenty of aleurioconidia. Aleurioconidia usually produced singly or in chains and lateral hyphae, brown, olivaceous, globose to subglobose, short stalks and obovate. The average sizes of conidia (n = 30) were 7.62~11.69 µm in length and 6.53~9.06 µm in width (Fig. 5I, 5J).
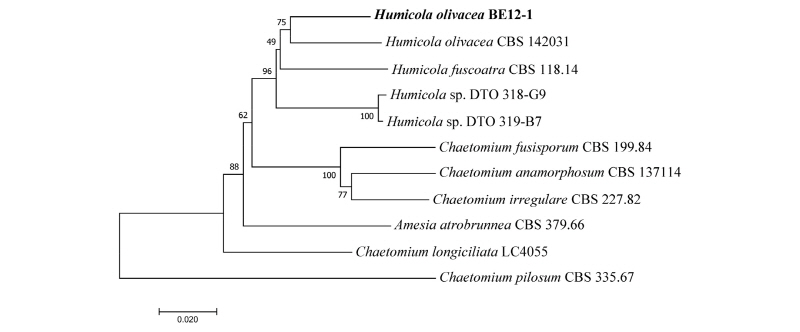
Fig. 6. Neighbor-joining phylogenetic tree based on a combined dataset of internal transcribed spacer region, large subunit, β-Tubulin, and RNA polymerase II genes sequences showing the relationships between Humicola olivacea and other members of the genus Humicola. The strain isolated in this study is indicated in bold. Bootstrap values are based on 1,000 replications. Bars, 0.02 substitutions per nucleotide position.
Molecular phylogeny of the isolate BE12-1
The phylogenetic relationship (Fig. 6) inferred from a combined dataset of ITS, LSU, β- Tubulin, RPB2 sequences revealed that the isolate BE12-1 was closely clustered with the Humicola olivacea strain (CBS 142031 or DTO 319-C7) with a high sequence similarity of 99%. The evolutionary relationship and phylogenetic analyses results support the identification of our isolate with H. olivacea.
The taxonomy of the genus Humicola is unclear, there are some infectious disease can cause by this genus [25] and also have numerous important applications in industries (like source of enzymes) [26]. To our knowledge, this is the first report of H. olivacea isolated from Korea.