Abstract
Rapid and accurate detection methods for Colletotrichum coccodes, an anthracnose pathogen of pepper and tomato, were developed using PCR. A specific primer set, coccoTef-F/coccoTef-R, which was constructed by analyzing tef-1α genes from 13 species and 22 strains of Colletotrichum, could specifically detect C. coccodes at a level of 10 ng by conventional PCR method and at 10 pg by real-time PCR. The PCR-based methods were also capable of detecting C. coccodes in pepper and tomato seeds artificially infected with the pathogen. The developed PCR methods can be applied for rapid and accurate inspection of C. coccodes in the seeds intended for export or import.
Acknowledgements
This study was supported by the Animal and Plant Quarantine Agency.
Figures & Tables
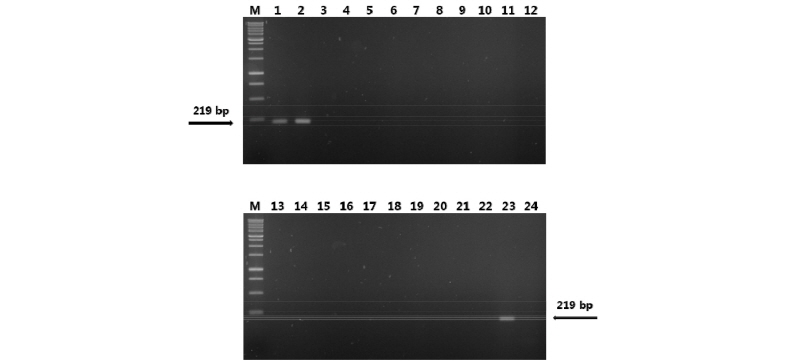
Fig. 1. Specificity test of the primer coccoTef-F/coccoTef-R against diverse fungal DNAs. Lane M, 1 kb DNA ladder; Lane 1, Colletotrichum coccodes KACC 40227; Lane 2, C. coccodes KACC 40802; Lane 3, C. orbiculare KACC 40808; Lane 4, C. orbiculare KACC 40903; Lane 5, C. circinans CBS 125331; Lane 6, C. circinans CBS 221.81; Lane 7~22, Other Colletotrichum spp. as listed in Table 1; Lane 23, DNA mixture of C. coccodes KACC 40802 and common six kinds of molds (Alternaria sp., Aspergillus sp., Cladosporium sp., Fusarium sp., Penicillium sp., Trichoderma sp.); Lane 24, DNA mixture of common six kinds of molds.


