INTRODUCTION
Freshwater is a diverse and complex environment for microorganisms, which provides several types of habitats for fungi such as plant litter including fallen leaves and decaying wood in freshwater, sediment, aquatic insects, and aquatic plants. To survive in a freshwater environment with limited nutrients and oxygen compared to the land environment, many fungi adhere to plant litter and/or sediment. Plant litter is one of the most important habitats for fungi, which function as decomposers [1]. Sediment is another well-known essential habitat for fungi that play a key role in biochemical processes in river ecosystems [2]. Freshwater fungi have been found in diverse habitats including plant litter and sediment, but their roles in aquatic ecosystems as well as their physiological and biochemical characteristics remain unclear.
Trichoderma Persoon is the largest genus in the family Hypocreaceae, containing 254 published names [3] and widely found in different climate zones [4]. Trichoderma species are typical cosmopolitan fungi which are isolated from diverse environments and substrates such as natural soils, decaying wood, agricultural habitats, mushroom, human body, and marine spaces [5, 6]. Most species are saprophytic fungi found on wood, bark, and other fungi, but some of species are associated with living plants as parasites, epiphytes, or endophytes [7]. Trichoderma species are among the most industrially useful fungi, but also are used as biocontrol agents to protect against plant disease or promote growth [8]. Despite the high species richness of Trichoderma, only 27 species have been reported in Korea and their habitats are limited to wood, soil, or mushrooms [9].
In this study, six Trichoderma species were isolated from plant litter and sediment in freshwater, T. afroharzianum, T. capillare, T. guizhouense, T. paraviridescens, T. reesei, and T. saturnisporum, which were not previously identified in Korea. We investigated the molecular phylogenetic and morphological characteristics of these species.
MATERIALS AND METHODS
Isolation of fungal strains and culture conditions
Fungal strains were collected from algae, plant litter, and soil sediment in a freshwater stream. Collection information of all strains used in this study is listed in Table 1. To isolate fungal strains from algae and plant litter, we washed the samples with distilled water at least twice, followed by incubation in pretreatment liquid medium (0.05% 3-morpholinopropane-1-sulfonic acid [weight/volume (w/v)], 0.05% KNO3 [w/v], 0.025% KH2PO4 [w/v], and 0.025% K2HPO4 [w/v]) at 20°C overnight. Then, 100 mL of the pretreatment medium was spread onto a 1% water agar plate and incubated at 20°C for 2 days. For soil sediments, we used a simple plating technique in which soil was distributed in a thin layer across a water agar plate. Hyphal tips and germinated conidia were isolated under a microscope and transferred onto a 24-well plate containing V8 agar (V8A; 8% V8 juice [v/v] and 1.5% agar [w/v] adjusted to pH 6.0 using 10N NaOH), and then incubated at 28°C in the dark. All strains used in this study were grown on potato dextrose agar (PDA; 3.9% potato dextrose agar powder [w/v]; Difco, Detroit, MI, USA), synthetic nitrogen-poor or nutrient-poor agar (SNA; 0.02% sucrose [w/v], 0.02% glucose [w/v], 0.1% KNO3 [w/v], 0.1% KH2PO4 [w/v], 0.05% MgSO4·7H2O [w/v], 0.05% NaCl [w/v], and 1.2% agar [w/v]), and corn meal dextrose agar (CMDA; 2% cornmeal [w/v], 2% glucose [w/v], and 2% agar). Mycelial growth patterns were recorded three days after inoculation at 28 °C on PDA, V8A, and corn meal agar (CMA; Difco, Detroit, MI, USA).
|
Table 1. Information of Trichoderma isolates isolated from freshwater environments 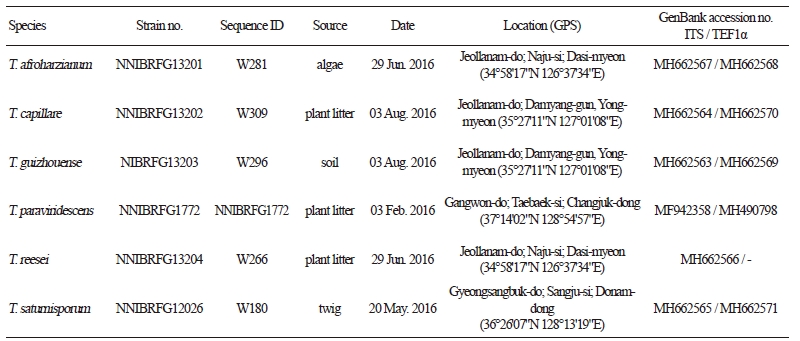
|
|
|
ITS, internal transcribed spacer; TEF1α, translation elongation factor 1. |
|
Extracellular enzyme activities
The enzyme activity of Trichoderma paraviridescens was detected as the formation of a clear zone around colonies on minimal malt extract agar (1% malt extract [w/v] and 1.5% agar [w/v]) with each substrate after 2 weeks of incubation at 25°C. Supplemented substrates were 0.5% soluble starch [w/v] for amylase activity, 0.5% tween 20 [w/v] for lipase activity, 0.5% carboxymethyl cellulose [w/v] for cellulase activity, and 1% skim milk [w/v] for protease activity in the presence of Congo red solution (500 ppm).
Morphological analysis
Conidiophores and conidia formed on three different media were transferred to a drop of distilled water on a slide glass and covered with a cover slip. Slides were examined and photographed using a BX53F microscope (Olympus, Tokyo, Japan) equipped with a DigiRetina 16M digital camera (Tucsen, Fuzhou, China) and an Eclipse Ni microscope (Nikon, Tokyo, Japan) equipped with a Ds-Ri2 digital camera (Nikon). At least 50 units were measured for each structure.
DNA extraction, PCR, and sequencing
Fungal genomic DNA was isolated with the NucleoSpin Plant II DNA extraction Kit (Macherey-Nagel, Düren, Germany) and MagListo 5M plant Genomic DNA Extraction Kit (Bioneer, Daejon, Korea). For molecular identification of the fungi, PCR amplification was performed for an internal transcribed spacer (ITS) rDNA region using primers ITS1 and ITS4 [10]. Translation elongation factor 1 (TEF1α) was amplified using two primer pairs: tef 85f/tef 954r [11] for the 5′ end and EF1-938F/EF1-1567R [12] for the 3′ end. Amplicons were sequenced by a DNA sequencing service (Macrogen, Seoul, Korea) with the same primers used for amplification. Similarity searching of the DNA sequences was carried out using BLAST algorithms available from the National Center for Biotechnology Information and ISTHinfo (www.isth.info, a website of Trichoderma DNA BarCode).
Phylogenetic analysis
ITS rDNA and the TEF1α sequences were edited using the DNASTAR software package version 5.05 (DNASTAR, Madison, WI, USA). Each locus was aligned using MAFFT 7 [13] and the Q-INS-i algorithm [14] with previously published sequences of the type or authentic isolates of Trichoderma species (listed in Table 2). Minimum Evolution (ME) method was used to construct phylogenetic trees. ME analysis was done using MEGA 7.0 software [15] with the default settings of the program, except for replacement with the Tamura-Nei model. Bootstrapping analysis of 1,000 replicates was performed to test the robustness of each grouping.
RESULTS AND DISCUSSION
Phylogenetic analysis
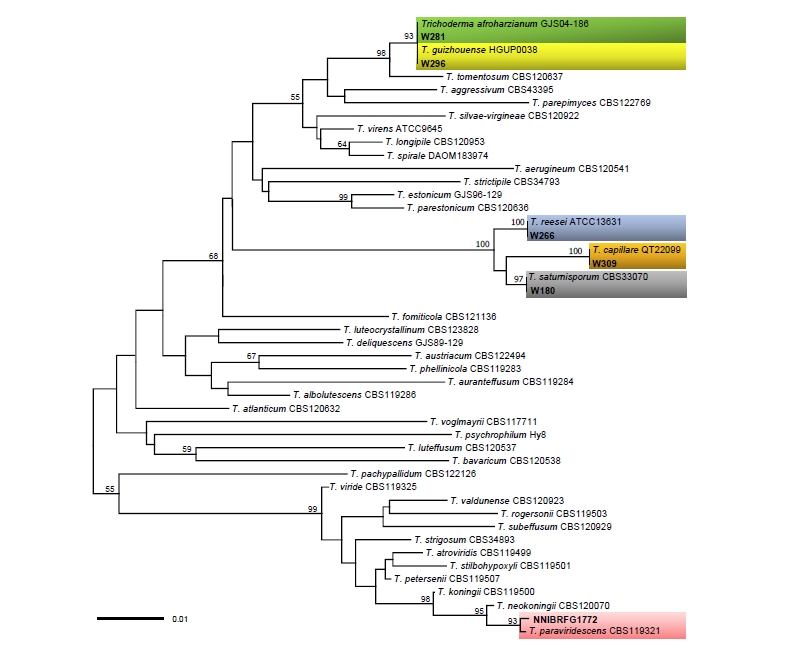
Sequences of two markers, ITS and TEF1α, were used to infer the phylogenetic relationships of Trichoderma species, including six Korean isolates and previously published authentic isolates. In ITS tree (Fig. 1), the accessions from the Korean isolates were placed into three distant clades. In the core of the “T. harzianum complex”, two isolates, W281 and W296, formed a clade with T. afroharzianum and T. guizhouense, showing no sequence differences. In TEF1α tree (Fig. 2A), however, the two isolates were clearly distinguished, each of which matched with T. afroharzianum and T. guizhouense. Three isolates, W180, W266, and W309, clustered in the second clade named as “the section Longibrachiatum” [16, 17] with maximum support, and then formed subgroups with T. saturnisporum, T. capillare, and T. reesei, respectively, with high supporting values of 100%, 100%, and 97%. Similarly, in the TEF1α tree, W180 and W309 formed two highly supported subgroups with T. saturnisporum and T. capillare, respectively. Isolate NNIBRFG1772 clustered with the type isolate CBS 119321 of T. paraviridescens in both trees (Fig. 1 and Fig. 2B) with high bootstrap values of 93% and 99%.
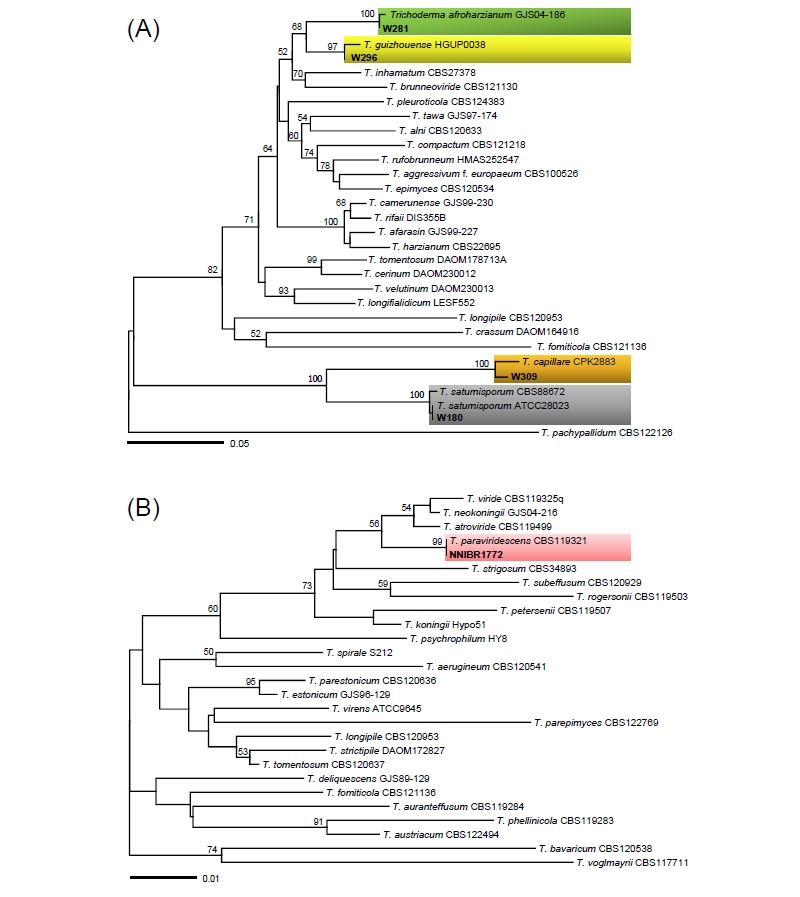
Fig. 2. Minimum evolution trees based on the translation elongation factor 1 (TEF1α) sequences; (A) for 5′ end of the TEF1α gene, (B) for 3′ end. Bootstrapping support values higher than 50% are shown above/below the branches. The specimens collected in Korea are shown in bold. The scale bar equals the number of nucleotide substitutions per site.
Extracellular enzyme activities of Trichoderma paraviridescens (NNIBRFG1772)
Many members of Trichoderma are used in wide range of industries including for enzyme production and as bio-control agents [8]. To screen for extracellular enzyme activities of T. paraviridescens, we performed growth tests using minimal malt extract agar media containing various substrates. After 14 days incubation at 25°C, enzyme activity was measured as the size and discoloration of the halo zone. Table 2 shows the extracellular enzyme activity including cellulase, amylase, lipase, and protease activities. As a result, T. paraviridescens (NNIBRFG1772) has high cellulase activity. In previous studies, many Trichoderma species were shown to be saprophytic fungi involved in decomposing biopolymers, and thus many studies of Trichoderma enzymes have focused on cellulase [18]. These results suggest that T. paraviridescens plays a role as a decomposer in freshwater ecosystems.
Taxonomy
Based on the molecular phylogenetic and morphological data, we identified six Trichoderma species previously unrecorded in Korea: T. afroharzianum, T. capillare, T. guizhouense, T. paraviridescens, T. reesei, and T. saturnisporum.
Trichoderma afroharzianum P. Chaverri, F.B. Rocha & I. Druzhinina, Mycologia 107 (3): 568 (2014) [MB#809945] (Fig. 3)
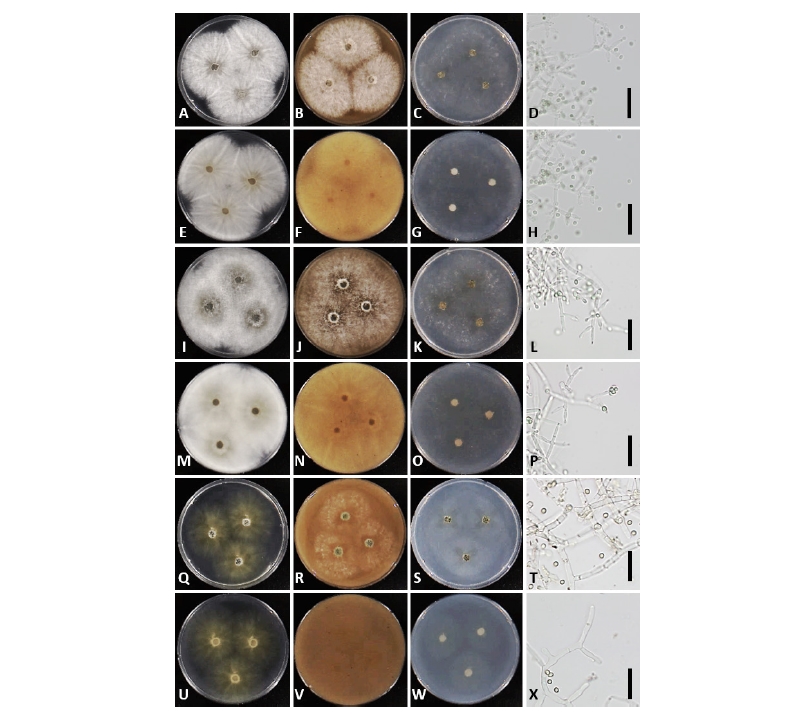
Fig. 3. Morphology of Trichoderma afroharzianum NNIBRFG13201 (W281) (A~H), Trichoderma capillare NNIBRFG13202 (W309) (I~P), and Trichoderma guizhouense NIBRFG13203 (W296) (Q~X). First row: colonies on potato dextrose agar (upper: observe view, lower: reverse view). Second row: colonies on V8 agar. Third row: colonies on corn meal agar. Fourth row: conidiophores and conidia (scale bars = 20 μm).
Description: Colonies on PDA and V8A at 28°C after 72 hr, 90 mm diameter, aerial mycelium white, abundant, dense; yellow pigment often diffused in the medium after 5 days. Colonies on CMA at 28°C after 72 hr, 75~80 mm diameter, aerial mycelium white, sparse. Conidiophores pyramidal, often symmetric, somewhat widely spaced branches, each branch terminating in a cruciate whorl or verticil of 3~5 phialides. Phialides lageniform to ampulliform, (3.5~)5.0~10.0(~16.5) × (2.0~)2.4~3.4(~4.0) µm, base (1.0~)1.5~2.0(~3.0) µm wide; supporting cell (2.0~)2.1~3.2(~4.5) µm wide. Conidia smooth, green to dark green, but rarely yellow, subglobose to ovoid, (2.0~)2.5~3.5(~4.5) × (2.0~)2.4~3.2(~3.5) µm. Chlamydospores not observed.
Isolate examined: Republic of Korea, Jeollanam-do; Naju-si; Dasi-myeon; Juksan-ri, in Juksan reservoir (34°58'17"N, 126°37'34"E), ex algae on water, June 29, 2016, Y.-J. Choi (NNIBRFG13201; W281)
Note: Trichoderma afroharzianum is a widespread species associated with diverse substrates, such as soils, roots, and other fungi, and thus is likely fungicolous [19]. Interestingly, the present collection was isolated from the surface of algae on freshwater. Since the epithet “afroharzianum” has been invalidly published by Druzhinina et al. [20], with no description, Chaverri et al. [21] has officially published this name. As shown previously [21], the Korean isolate exhibited long phialides and narrow supporting cell for the phialides, but also grew well even at 35°C.
Trichoderma capillare Samuels & C.P. Kubicek, Fungal Diversity 55: 83 (2012) [MB#563903] (Fig. 3)
Description: Colonies on PDA and V8A at 28°C after 72 hr, 80 mm diameter, aerial mycelium white, abundant, dense; yellow to brown pigment often diffused in the medium after 96 hr. Colonies on CMA at 28°C after 72 hr, 75~80 mm diameter, aerial mycelium white, fluffy, sparse. Conidiophores highly variable in form; secondary branches commonly producing solitary phialides, with intercalary phialides, but also often terminating in whorls of a few phialides, and rarely lacking any obvious pattern, with cells of fertile branches sometimes vesiculose and producing numerous phialides. Phialides lageniform, mostly straight, nearly cylindrical or conspicuously swollen below the middle, (4.5~)7.0~10.0(~15.0) µm long, (2.0~)3.0~3.5(~4.0) µm at the widest point, (1.0~)2.2~3.2 µm wide at the base. Conidia green, roughened, less frequently smooth, subglobose to broadly ellipsoidal, (2.0~)2.5~4.0(~5.0) × (1.5~)2.2~3.3(~4.0) µm. Chlamydospores not observed.
Isolate examined: Republic of Korea, Jeollanam-do; Damyang-gun; Yong-myeon, at Gamagol Ecological Park (35°27'11"N 127°01'08"E), ex a decaying leaf in freshwater, August 3, 2016, Y.-J. Choi (NNIBRFG13202; W309)
Note: This species is known in Europe, USA, Colombia, Vietnam, and Taiwan. This is the first report of T. capillare in Korea. The morphological characteristics of Korean species are consistent with the descriptions of Samuels et al. [22]. Trichoderma capillare exhibits an unusual branching pattern and phialides arrangement, which were previously marked as key characters in the identification of this species [7, 23].
Trichoderma guizhouense Q.R. Li, E.H.C McKenzie & Yong Wang, Mycological Progress 12: 170 (2013) [MB#563664] (Fig. 3)
Description: Colonies on PDA at 28°C after 72 hr, 70 mm diameter, aerial mycelium white, dense; yellow to brownish pigment often diffused in the medium after 96 hr. Colonies on V8A at 28°C after 72 hr, 55 mm diameter, aerial mycelium white, abundant, sparse; yellow pigment often diffused in the medium after 5 days. Colonies on CMA after 72 hr at 28°C, 25 mm diameter, aerial mycelium white, very lightly. Conidiophores pyramidal, often symmetric, somewhat widely spaced branches, each branch terminating in a cruciate whorl of 2~4 phialides. Phialides ampulliform to lageniform, typically strongly constricted below the tip, (3.5~)4.5~8.0(~11.5) × (2.0~)2.8~3.5(~4.0) µm, base (1.0~)1.5~2.5(~3.0) µm wide. Conidia smooth, green, subglobose to ovoid, (2.0~)2.5~3.5(~4.0) × (2.0~)2.5~3.0(~3.5) µm. Chlamydospores not observed.
Isolate examined: Republic of Korea, Jeollanam-do; Damyang-gun; Yong- myeon, at Gamagol Ecological Park (35°27'11"N 127°01'08"E), ex under freshwater, August 3, 2016, Y.-J. Choi (NNIBRFG13203; W296)
Note: Trichoderma guizhouense is distributed worldwide. This species can be divided into two groups with endophytic vs. non-endophytic habitats [21]. As the Korean isolate was isolated from a decaying leaf on the bottom of a freshwater stream, it is likely that the Korean isolate is close to the entophytic group of this species, but the morphological differences observed between the two groups [21] were unclear in the present isolate. Trichoderma guizhouense is phylogenetically close to T. afroharzianum in the complex, but its slower growth on PDA differs from the latter species.
Trichoderma paraviridescens Jaklitsch, Samuels & Voglmayr Persoonia 31: 128 (2013) [MB#803623] (Fig. 4)
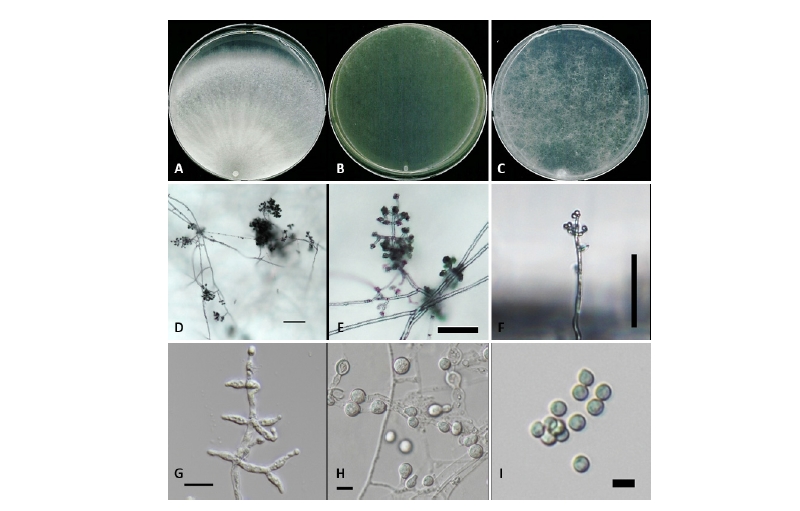
Fig. 4. Trichoderma paraviridescens NNIBRFG1772. Colony morphology on potato dextrose agar (A), corn meal dextrose agar (B) and synthetic nitrogen-poor or nutrient-poor agar (C) after 7 days at 25℃. D~G, Conidiophore; H, Chlamydospores; I, Conidia (scale bars: D, E = 100 μm, F = 50 μm, G~I = 10 μm).
Description: Colonies on PDA after 7 days 70~75 mm diameter at 25°C, white abundant and dense aerial mycelium. Colonies on CMDA after 7 days 80~85 mm diameter at 25°C, feathery aerial hyphae, white to yellow green. Colonies on SNA after 7 days 75~80 mm diameter at 25°C, downy aerial hyphae, hyaline to white. Conidiation noted after 2~3 days, turn to green after 4~5 days. Conidiophores tree/branch-like from, consist of a distinct main axis with 1.3~3.8 µm wide side branches, re-branching to form second branches. Some conidiophores broad and dense, with mostly symmetric branches. Phialides (5.6~)7.6~9.6(~12.3) × (2.0~)2.7~3.0(~3.4) µm, l/w ratio (1.6~)2.4~3.7(~4.3), base (1.3~)1.5~2.1(~2.7) µm wide (n = 25), lageniform, sometimes divergent in small whorls of 2~3. Conidia globose to subglobose, 2.3~4.9 × 1.82~3.51 µm (av. 3.30 ± 0.54 × 2.67 ± 0.49 µm; n = 28), l/w ratio av. 1.27 ± 0.30 (n = 28), green. Chlamydospores observed after 2~3 days at 25°C; numerous on entire plate, 6.08~11.17 (av. 8.91 ± 1.50) µm in diameter (n = 25), globose, with thick wall.
Isolate examined: Republic of Korea, Gangwon-do; Taebaek-si; Changjuk-dong (37°14'02"N 128°54'57"E), ex plant litter deposited in the stream Geomryongso, February 3, 2016, J. Goh, (NNIBRFG1772)
Note: The type isolate CBS 119321 of T. paraviridescens has been isolated from decorticated branch of Fagus sylvatica [24]. The present isolate was isolated from plant litter in freshwater, adding it to a new habitat of T. paraviridescens. In addition, the present study for extracellular enzyme activities revealed that T. paraviridescens produces various organic matters, which has a high potential for enhancing highly fungal biodiversity in Korea.
Trichoderma reesei E.G. Simmons, Abstracts, 2nd International Mycological Congress: 618 (1977) [MB#324906] (Fig. 5)
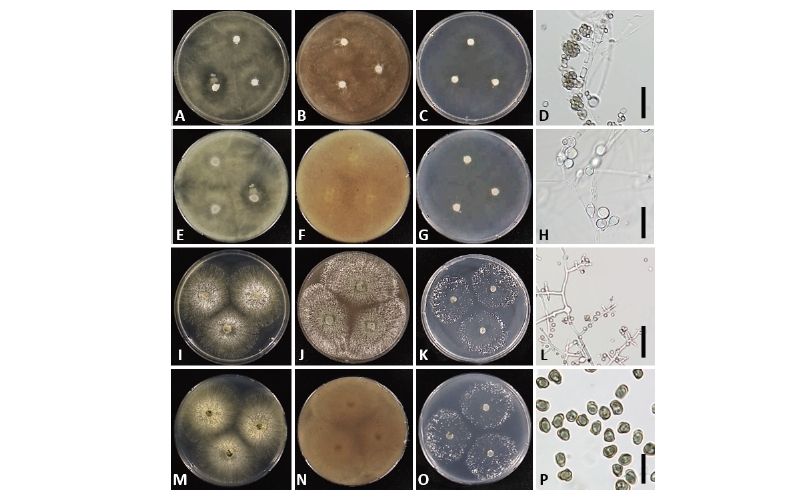
Fig. 5. Morphology of Trichoderma reesei NNIBRFG13204 (W266) (A~H) and T. saturnisporum NNIBRFG12026 (W180) (I~P). First row: colonies on potato dextrose agar (upper: observed view, lower: reverse view). Second row: colonies on V8 agar. Third row: colonies on corn meal agar. Fourth row: conidiophores and conidia. Chlamydospores of T. reesei (H) (scale bars: D, H, L = 20 μm, P = 10 μm).
Teleomorph: Hypocrea jecorina Berk. & Broome, J. Linn. Soc. Bot. 14: 112 (1873).
Ex-type culture: ATCC 13631 = CBS 383.78
Description: Colonies grew rapidly on PDA and V8A completely, filling the petri plate of 90 mm diameter after 72 hr at 28°C, white, aerial mycelium. Colonies on CMA at 25°C after 72 hr. Approximately 50 mm, white, less dense and lightly. Conidiophores comprising a long central axis with 5~8 levels of solitary phialides before commencement of secondary branching; secondary branches often comprising a single cell terminated by a single phialide or up to 4 cells and single cells terminated by a solitary phialide toward the base at the main axis. Phialides lageniform, asymmetric, swollen or not at the middle, straight, less frequently sinuous, (3.0~)6.0~9.0(~11.5) µm long, (1.8~)2.3~3.0(~4.0) µm at the widest point, base (1.0~)1.3~2.7(~3.0) µm. Conidia ellipsoidal to oblong, green, smooth, (3.0~)3.5~4.5(~6.0) × (1.5~)2.5~3.0(~3.5) µm. Chlamydospores hyaline, subglobose to pyriform, mainly terminal.
Isolate examined: Republic of Korea, Jeollanam-do; Naju-si; Dasi-myeon; Juksan-ri, in Juksan reservoir (34°58'17"N 126°37'34"E), ex decaying herbaceous plants under water, June 29, 2016, Y.-J. Choi (NNIBRFG13204; W266)
Note: This is the first report of T. reesei, a cosmopolitan and tropical species, from South Korea in a subtropical area. Trichoderma reesei is one of the most economically important species in the genus because of its extraordinary ability to produce cellulolytic and hemicellulolytic enzymes used for hydrolysis of lignocelluloses in the food and feed industries [22, 25]. Trichoderma reesei and T. capillare belong to the clade Longibrachiatum, but the former species formed relatively rare aerial mycelium on all media of PDA, V8A, and CMA.
Trichoderma saturnisporum Hammill, Mycologia 62(1): 112 (1970) [MB#324907] (Fig. 5)
Description: Colonies on PDA and V8A at 28°C after 72 hr, 70 mm diameter, aerial mycelium white, abundant, dense; uncolored or yellowish pigment often diffused in the medium after 96 hr. Colonies on CMA at 28°C after 72 hr, 50~70 mm diameter, aerial mycelium white, fluffy, sparse. Conidiophores highly variable in form; secondary branches commonly producing solitary or alternate phialides, but also often terminating in whorls of paired phialides. Phialides various from mostly lageniform to ampulliform, often curved, (5.5~)7.0~12.0(~14.0) µm long, (2.0~)2.5~3.0(~3.5) µm at the widest point, (1.0~)2.0~3.0 µm wide at the base. Conidia subglobose to ellipsoidal or sub-cylindrical, dark green, with wart-like ornamentations, but less frequently smooth, (3.0~)3.5~5.0(~6.0) × (2.0~)2.5~3.0(~4.0) µm. Chlamydospores not observed.
Isolate examined: Republic of Korea, Gyeongsangbuk-do; Sangju-si; Donam-dong, in Sangju reservoir (36°26'07"N 128°13'19"E), ex a decaying wooden twig under water, May 20, 2016, Y.-J. Choi (NNIBRFG12026; W180)
Note: Trichoderma saturnisporum is a cosmopolitan species, which is widely distributed in North America, Caribbean Ocean region, Europe, South Africa, Australia [22], and Asia (China) [16]. This is the first report of this species in Korea. Because of the warts or tubercules on its conidia, T. saturnisporum is easily distinguished from other species of this genus [26]. Interestingly, it was shown that T. saturnisporum improves plant growth, but also exhibits biocontrol activity against infections by Phytophthora spp. [27].