INTRODUCTION
The genus Nannizziopsis is a member of the family Nannizziopsiaceae, the order Onygenales, and can be obtained from soil. Fungal species belonging to the Nannizziopsiaceae have the peculiar character of causing skin infections in reptiles [1]. In addition, the members of this family produce hyaline, thin, and one-celled conidia [1]. This family lies under the largest phylum of the kingdom Fungi, Ascomycota. There are more than 64,000 species of ascomycetes [2]. The typical feature of the ascomycetes is the presence of the ascus, a microscopic sexual structure containing nonmotile spores called ascospores [2].
The genus Nannizziopsis was first described by Currah [3] in 1985 to represent a single species, Rollandina vriesii Apinis [4]. Apinis’ idea to the genus Rollandina was initially based on the structure of hyphae found on the stalk [4], since this structure appears to be based upon two fungi, the stalk, stipe of a small mushroom, and the ascomata of a Nannizzia and its Microsporum anamorph [5]. According to Uchuyama et al. [6], Currah [3] had proposed the new genus Nannizziopsis and proposed the new combination. This genus is characterized by the presence of white ascomata, peridial asperulate hyphae constricted at the septa, and hyaline and globose ascospores [6].
During a survey of a Korean indigenous fungal species from field soils, Nannizziopsis mirabilis was isolated from Jeju-do field soil in 2017. This fungal species has not been officially reported before, and herein, we are reporting this species as a new record from Korea. The prime objective of this study was to: (1) study the morphology and molecular aspects of N. mirabilis, and (2) to compare and distinguish this newly recorded species with previously reported N. mirabilis species.
MATERIALS AND METHODS
Soil sample collection and isolation of fungi
Soil samples were brought from agricultural field soils in Jeju-do, Korea. The samples were collected from a depth of 0~15 cm, air dried, and stored in plastic bags at 4°C until use. Approximately 200~250 g of soil was collected for each sample. Fungal isolates were isolated from a conventional dilution plating technique [7]. One gram of the soil sample was suspended in nine milliliters of sterile distilled water; the mixture was shaken vigorously for 2~3 min. The soil suspensions were diluted serially in sterile distilled water. One milliliter each from the dilutions, 10-3, 10-4, 10-5 and 10-6, was pipetted and streaked onto petri plates containing potato dextrose agar (PDA; Difco, Detroit, MI, USA) supplemented with 100 μg/L chloramphenicol (a bacteriostatic agent) for 5~7 days at 25°C until the growth of fungal colonies was observed. The pure cultures were maintained on PDA slants at 4°C for future use.
Morphological description
Morphological characteristics of isolates KNU17-67 was observed on PDA (MB Cell, Los Angeles, CA, USA), at 25°C in the dark for 7 days. Medium was prepared as described by Samson et al. [8]. After incubation, the diameter of the colonies was measured, and the degree of sporulation was determined. Colony color (obverse and reverse side) was described using a Methuen handbook of color [9]. Microscopic pictures were taken using an HK 3.1 CMOS digital camera (KOPTIC, Seoul, Korea) attached to an Olympus BX50F-3 microscope (Olympus, Tokyo, Japan) and a scanning electron microscope (LEO Model 1450VP Variable Pressure Scanning Electron Microscope; Carl Zeiss, Oberkochen, Germany). Detailed morphological features of the isolate have been presented in Table 1.
|
Table 2. List of ITS rDNA sequences of Nannizziopsis mirabilis and its allied species used in this study, along with their isolate and GenBank accession numbers 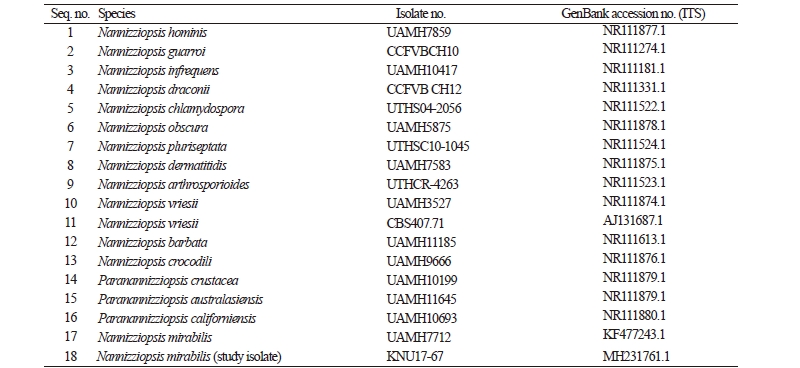
|
|
|
ITS, internal transcribed spacer. |
|
Genomic DNA extraction, sequencing, and data analysis
Total genomic DNA was extracted from isolate KNU17-67 using a DNeasy Plant Mini kit (Qiagen, Germantown, MD, USA), following the manufacturer’s instructions. The internal transcribed spacer (ITS) region of rDNA was amplified by the primers ITS1 (5′-TCCGTAGGTGAACCTGCG-3′) and ITS4 (5′-TCCTCCGCTTATTGATATGC-3′) [10]. The amplified PCR products were sequenced using an ABI Prism 3730 DNA analyzer (Applied Biosystems, Foster City, CA, USA). The sequences were compared with reference ITS sequences from GenBank at the National Center for Biotechnology Information (NCBI), using the basic local alignment search tool [11]. The nucleotide sequence of KNU17-67 was deposited at the culture collection facility of the National Institute of Biological Resources (NIBR, NIBRFG0000501850). The nucleotide sequences were also deposited in GenBank and assigned the accession number MH231761. The isolates used in this study to construct the phylogenetic tree were summarized in Table 2 with their strain and GenBank accession numbers. A phylogenetic relationship was analyzed using molecular evolutionary genetic analysis (MEGA 6) software [12]. A neighbor-joining tree was constructed using the Kimura two-parameter substitution model [13]. Bootstrap analysis was performed with 1,000 replications to determine the support for each clade.
RESULTS AND DISCUSSION
Nannizziopsis mirabilis Uchy, Kamiya and Udagawa. Mycoscience 36 (2): 205 (1995). [MB#413562].
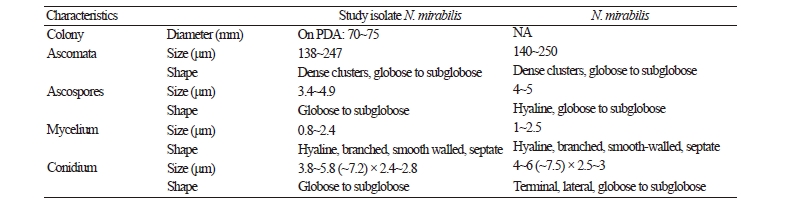
Morphology of the isolate KNU17-67
Macro and micro morphological pictures of the isolate are shown in Fig. 1. Colony growth on PDA was fast; the colonies attained a diameter of 70~75 mm after growth for 7 days at 25°C. The front and back side of the colony was white and yellow color, respectively (Fig 1A, 1B). The texture of the colony was floccose; the sporulation was dense (Fig. 1C, 1F). The form of the colony was irregular, and the surface was smooth. Ascomata were dense and in clusters, globose to subglobose, and 138~247 μm in diameter. Mycelium was hyaline, branched, smooth-walled, septate, and 0.8~2.4 μm diameter in size. Ascopores were found to be globose to subglobose in shape and have a diameter of 3.4~4.9 μm (Fig. 1D, 1E). Conidia were globose and subglobose, with a size of 3.8~5.8 (~7.2) × 2.4~2.8 μm (Fig. 1D). The morphological characters of the isolate N. mirabilis were compared with those of the previously reported N. mirabilis [6]. The shape and size of the ascospores and conidia of the studied isolate fit reasonably with the description of N. mirabilis reported by Uchuyama et al. [6], except for the subtle differences in the sizes of the ascomata, ascospores, and conidia. However, the similar macro and micromorphological characters of the studied isolate suggest that the isolate is N. mirabilis.
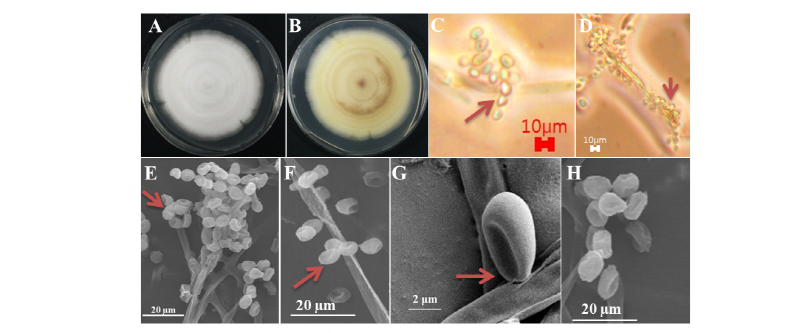
Fig. 1. Morphological characters of Nannizziopsis mirabilis KNU17-67 grown on potato dextrose agar for 7 days at 25°C. A, Front colony; B, Back colony; C, Simple microscopic picture of conidia; D, Simple microscopic picture of apical conidia; E, Scanning electron microscopic picture of terminal conidia; F, Scanning electron microscopic picture of apical and terminal conidia; G, H, Scanning electron microscopic picture of ascospores.
Molecular phylogeny of the isolate KNU17-67
To study the phylogenetic relationship between the studied isolate and previously reported N. mirabilis, their ITS rDNA sequences were compared and analyzed. As shown in Fig. 2, the study isolate was clustered in the same group, with 100% similarity with N. mirabilis.
Based on both morphological and molecular characters, the studied isolate KNU17-67 was identified as N. mirabilis, which is new in Korea. It has some valuable biotechnological importance that would be valuable in the field of fungal taxonomy and fungal biotechnology. Conclusively, further studies on the isolation and identification of several new fungal species within the Ascomycota kingdom are required to extend our knowledge about the indigenous fungal species in Korea.